Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
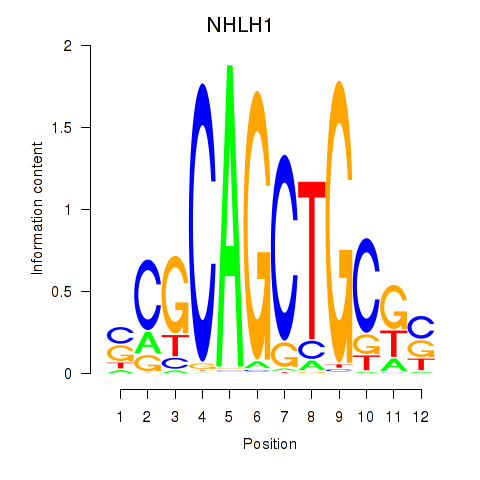
Results for NHLH1
Z-value: 1.25
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | 0.65 | 3.5e-01 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_7005785 | 1.43 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr11_-_64764435 | 0.91 |
ENST00000534177.1
ENST00000301887.4 |
BATF2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr14_-_37051798 | 0.77 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr4_+_89299994 | 0.76 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr4_+_89299885 | 0.68 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_+_35521572 | 0.64 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr3_-_45187843 | 0.63 |
ENST00000296129.1
ENST00000425231.2 |
CDCP1
|
CUB domain containing protein 1 |
| chr16_+_77246337 | 0.56 |
ENST00000563157.1
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_+_35521616 | 0.55 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr6_-_134861089 | 0.53 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr20_-_23669590 | 0.52 |
ENST00000217423.3
|
CST4
|
cystatin S |
| chr6_+_126112001 | 0.52 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr15_+_90735145 | 0.49 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr7_-_100808843 | 0.47 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr17_-_54991369 | 0.46 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chr19_-_40730820 | 0.46 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr10_-_101380121 | 0.45 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr5_+_49962772 | 0.45 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_+_196522032 | 0.45 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr10_+_102758105 | 0.44 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr17_+_38171614 | 0.43 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr16_-_89268070 | 0.41 |
ENST00000562855.2
|
SLC22A31
|
solute carrier family 22, member 31 |
| chr4_-_80994210 | 0.40 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr14_-_101053739 | 0.39 |
ENST00000554140.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr19_+_1286097 | 0.39 |
ENST00000215368.2
|
EFNA2
|
ephrin-A2 |
| chr1_+_113217309 | 0.38 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr7_+_130794846 | 0.37 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr1_-_47184745 | 0.37 |
ENST00000544071.1
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr6_-_38607673 | 0.37 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr19_+_35521699 | 0.36 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chrX_-_47004437 | 0.36 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr17_+_74261413 | 0.35 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr11_-_6341724 | 0.35 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr6_-_43596899 | 0.35 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr19_+_4304685 | 0.34 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr2_+_8822113 | 0.34 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr14_+_64971438 | 0.33 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr12_-_54779511 | 0.33 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr17_-_54991395 | 0.33 |
ENST00000316881.4
|
TRIM25
|
tripartite motif containing 25 |
| chr17_+_80193644 | 0.32 |
ENST00000582946.1
|
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_-_94620239 | 0.32 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr12_+_122064673 | 0.31 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr19_-_17445613 | 0.31 |
ENST00000159087.4
|
ANO8
|
anoctamin 8 |
| chr20_-_31172598 | 0.30 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr20_+_49348109 | 0.30 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr1_+_161692449 | 0.30 |
ENST00000367946.3
ENST00000367945.1 ENST00000336830.5 ENST00000367944.3 ENST00000392158.1 |
FCRLB
|
Fc receptor-like B |
| chr20_-_35274548 | 0.29 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr19_+_41107249 | 0.29 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_+_39382900 | 0.29 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr7_-_100808394 | 0.29 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr11_-_10830463 | 0.29 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr11_-_6341844 | 0.29 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr2_-_175202151 | 0.28 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr3_-_5229982 | 0.28 |
ENST00000600805.1
|
AC026202.1
|
Uncharacterized protein |
| chr12_-_113658826 | 0.28 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr9_-_131940526 | 0.27 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr16_+_330581 | 0.27 |
ENST00000219409.3
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr4_+_75311019 | 0.27 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr6_-_160148356 | 0.27 |
ENST00000401980.3
ENST00000545162.1 |
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr19_+_39989580 | 0.26 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr15_-_56209306 | 0.26 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr19_-_38743878 | 0.26 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr12_-_54813229 | 0.26 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chrX_-_45710920 | 0.25 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr14_+_102027688 | 0.25 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr2_-_232791038 | 0.25 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr7_-_138720763 | 0.25 |
ENST00000275766.1
|
ZC3HAV1L
|
zinc finger CCCH-type, antiviral 1-like |
| chr6_+_145118873 | 0.25 |
ENST00000432686.1
ENST00000417142.1 |
UTRN
|
utrophin |
| chr22_-_50964849 | 0.25 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr4_+_75310851 | 0.25 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr6_+_31895467 | 0.25 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr12_-_74686314 | 0.25 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr22_-_36850991 | 0.25 |
ENST00000442579.1
|
RP5-1119A7.14
|
RP5-1119A7.14 |
| chr9_-_100459639 | 0.25 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr6_+_31895480 | 0.24 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_+_113217345 | 0.24 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr8_+_86376081 | 0.24 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr22_+_19705928 | 0.24 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr19_-_40732594 | 0.24 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr1_-_33336414 | 0.24 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr12_-_72057638 | 0.24 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr20_-_48770244 | 0.23 |
ENST00000371650.5
ENST00000371652.4 ENST00000557021.1 |
TMEM189
|
transmembrane protein 189 |
| chr17_+_48638371 | 0.23 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr19_-_47104118 | 0.23 |
ENST00000593888.1
ENST00000602017.1 |
AC011551.3
PPP5D1
|
Uncharacterized protein PPP5 tetratricopeptide repeat domain containing 1 |
| chr2_-_102003987 | 0.23 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr14_+_90863364 | 0.23 |
ENST00000447653.3
ENST00000553542.1 |
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr4_-_54930790 | 0.23 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr19_+_13228917 | 0.23 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_+_7338737 | 0.23 |
ENST00000323206.1
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr4_-_41216619 | 0.23 |
ENST00000508676.1
ENST00000506352.1 ENST00000295974.8 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr17_-_31620006 | 0.22 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr1_-_244013384 | 0.22 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr1_-_197744763 | 0.22 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr17_-_71228357 | 0.22 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr4_+_153457404 | 0.22 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr1_+_150039877 | 0.22 |
ENST00000419023.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr2_+_58655461 | 0.22 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr20_+_30193083 | 0.22 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr5_-_95297534 | 0.22 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr18_-_51750948 | 0.22 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr19_-_19051103 | 0.22 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr17_-_27278445 | 0.21 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr12_-_72057571 | 0.21 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr9_-_4679419 | 0.21 |
ENST00000609131.1
ENST00000607997.1 |
RP11-6J24.6
|
RP11-6J24.6 |
| chr2_+_10442993 | 0.21 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr6_+_146056706 | 0.20 |
ENST00000603994.1
|
RP3-466P17.1
|
RP3-466P17.1 |
| chr7_-_100171270 | 0.20 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr11_-_10829851 | 0.20 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr17_-_73401728 | 0.20 |
ENST00000316804.5
ENST00000316615.5 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr1_-_226065330 | 0.20 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr12_-_113658892 | 0.20 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr20_-_48770174 | 0.20 |
ENST00000341698.2
|
TMEM189-UBE2V1
|
TMEM189-UBE2V1 readthrough |
| chr5_-_169816638 | 0.20 |
ENST00000521859.1
ENST00000274629.4 |
KCNMB1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr3_-_133614597 | 0.19 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr9_-_127905736 | 0.19 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr9_+_101569944 | 0.19 |
ENST00000375011.3
|
GALNT12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
| chr12_+_57984965 | 0.19 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr16_+_1756162 | 0.19 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr17_-_16395455 | 0.19 |
ENST00000409083.3
|
FAM211A
|
family with sequence similarity 211, member A |
| chr7_+_72848092 | 0.19 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr15_+_74833518 | 0.19 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chrX_-_19905703 | 0.19 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_+_74261277 | 0.19 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr1_-_91487806 | 0.19 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chrX_-_84634737 | 0.19 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr17_-_56606639 | 0.19 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr20_-_30433396 | 0.19 |
ENST00000375978.3
|
FOXS1
|
forkhead box S1 |
| chr13_+_49550015 | 0.19 |
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr9_+_110045418 | 0.19 |
ENST00000419616.1
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr1_-_47134085 | 0.19 |
ENST00000371937.4
ENST00000574428.1 ENST00000329231.4 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr18_+_9913977 | 0.19 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr21_-_46293586 | 0.18 |
ENST00000445724.2
ENST00000397887.3 |
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein |
| chr1_+_150039787 | 0.18 |
ENST00000535106.1
|
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr17_-_31204124 | 0.18 |
ENST00000579584.1
ENST00000318217.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr2_+_23608064 | 0.18 |
ENST00000486442.1
|
KLHL29
|
kelch-like family member 29 |
| chr3_+_159481988 | 0.18 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr5_+_176873446 | 0.18 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr1_-_85155939 | 0.18 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr12_+_52404270 | 0.18 |
ENST00000552049.1
ENST00000546756.1 |
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr2_-_178937478 | 0.18 |
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A |
| chr20_+_49348081 | 0.18 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr12_-_6579808 | 0.18 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr5_-_59064458 | 0.18 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_-_64512273 | 0.18 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_-_92351666 | 0.17 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr2_+_233734994 | 0.17 |
ENST00000331342.2
|
C2orf82
|
chromosome 2 open reading frame 82 |
| chr2_+_7005959 | 0.17 |
ENST00000442639.1
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr10_-_27529779 | 0.17 |
ENST00000426079.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr3_-_172428959 | 0.17 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr8_-_98290087 | 0.17 |
ENST00000322128.3
|
TSPYL5
|
TSPY-like 5 |
| chr7_-_77045617 | 0.17 |
ENST00000257626.7
|
GSAP
|
gamma-secretase activating protein |
| chr14_-_64971288 | 0.17 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr17_-_56606705 | 0.17 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr3_+_96533621 | 0.17 |
ENST00000542517.1
ENST00000506569.1 |
EPHA6
|
EPH receptor A6 |
| chr19_+_39989535 | 0.17 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr2_-_233352531 | 0.17 |
ENST00000304546.1
|
ECEL1
|
endothelin converting enzyme-like 1 |
| chr7_-_105925367 | 0.17 |
ENST00000354289.4
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr13_-_26625169 | 0.17 |
ENST00000319420.3
|
SHISA2
|
shisa family member 2 |
| chr19_+_17416609 | 0.17 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr19_-_49944806 | 0.17 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr17_+_4981535 | 0.17 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr21_+_38071430 | 0.17 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr3_-_68981685 | 0.16 |
ENST00000495737.1
ENST00000295569.7 |
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 |
| chr5_+_176873789 | 0.16 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr14_-_24584138 | 0.16 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr1_-_52344416 | 0.16 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr21_+_34442439 | 0.16 |
ENST00000382348.1
ENST00000333063.5 |
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr12_-_118406777 | 0.16 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr16_+_11439286 | 0.16 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr7_+_7811992 | 0.16 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr19_+_46001697 | 0.16 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr10_+_105036909 | 0.16 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr4_-_168155730 | 0.16 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_+_196521845 | 0.16 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr17_-_56606664 | 0.16 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr2_+_16080659 | 0.16 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr2_+_47168313 | 0.15 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr11_+_62556596 | 0.15 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr3_-_78719376 | 0.15 |
ENST00000495961.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_45737469 | 0.15 |
ENST00000413988.1
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr2_-_73511407 | 0.15 |
ENST00000520530.2
|
FBXO41
|
F-box protein 41 |
| chr1_-_19536744 | 0.15 |
ENST00000375267.2
ENST00000375217.2 ENST00000375226.2 ENST00000375254.3 |
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr1_+_157963063 | 0.15 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr17_-_79792909 | 0.15 |
ENST00000330261.4
ENST00000570394.1 |
PPP1R27
|
protein phosphatase 1, regulatory subunit 27 |
| chr1_-_11107280 | 0.15 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr1_+_20915409 | 0.15 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr6_-_30712313 | 0.15 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr1_+_39456895 | 0.15 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr19_+_1941117 | 0.15 |
ENST00000255641.8
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chrX_+_107069063 | 0.15 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr17_+_66255310 | 0.15 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr16_+_3014269 | 0.14 |
ENST00000575885.1
ENST00000571007.1 ENST00000319500.6 |
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr17_-_73401567 | 0.14 |
ENST00000392562.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr6_-_75915757 | 0.14 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr1_+_161185032 | 0.14 |
ENST00000367992.3
ENST00000289902.1 |
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr2_+_191045656 | 0.14 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr1_+_27113963 | 0.14 |
ENST00000430292.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chrX_-_134049262 | 0.14 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr8_-_17270809 | 0.14 |
ENST00000180173.5
ENST00000521857.1 |
MTMR7
|
myotubularin related protein 7 |
| chr12_+_121088291 | 0.14 |
ENST00000351200.2
|
CABP1
|
calcium binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.4 | 1.4 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.4 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.2 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.1 | 0.5 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.4 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.5 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.2 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.2 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.5 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.6 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.3 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0033024 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) positive regulation of mast cell cytokine production(GO:0032765) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:0032804 | viral protein processing(GO:0019082) negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.3 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 1.0 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.9 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.3 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.2 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.0 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.8 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.3 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0060503 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.0 | GO:0034699 | response to luteinizing hormone(GO:0034699) |
| 0.0 | 0.3 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.5 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.0 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.4 | 1.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.2 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.1 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |