Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
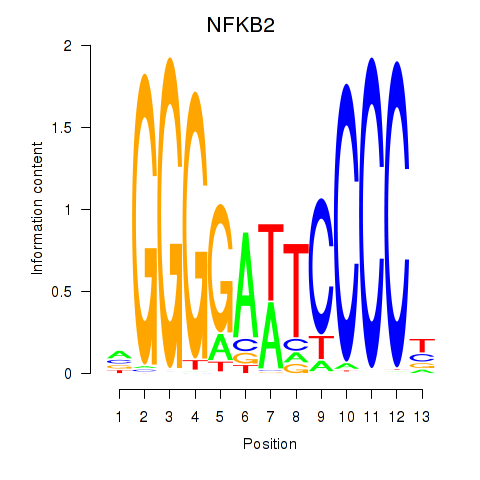
Results for NFKB2
Z-value: 1.04
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.13 | nuclear factor kappa B subunit 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg19_v2_chr10_+_104155450_104155479 | 0.82 | 1.8e-01 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34207295 | 1.40 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr11_-_57335750 | 0.92 |
ENST00000340573.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr14_-_24616426 | 0.83 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr12_-_56753858 | 0.78 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr19_-_55881741 | 0.69 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr11_+_60691924 | 0.65 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr15_-_89438742 | 0.59 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr5_-_179780312 | 0.59 |
ENST00000253778.8
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr6_-_31324943 | 0.59 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr4_+_8271471 | 0.58 |
ENST00000307358.2
ENST00000382512.3 |
HTRA3
|
HtrA serine peptidase 3 |
| chr16_-_74734742 | 0.56 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr16_+_67280799 | 0.52 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr11_-_33891362 | 0.49 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr10_+_104154229 | 0.48 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_+_176972000 | 0.47 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr4_-_74864386 | 0.46 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr18_+_21529811 | 0.45 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr22_-_20255212 | 0.43 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr14_-_24615805 | 0.43 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr1_-_41328018 | 0.42 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr19_+_45504688 | 0.41 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr20_+_43803517 | 0.41 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr7_+_1570322 | 0.40 |
ENST00000343242.4
|
MAFK
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr17_-_56605341 | 0.39 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr11_-_18270182 | 0.39 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr13_-_41111323 | 0.39 |
ENST00000595486.1
|
AL133318.1
|
Uncharacterized protein |
| chr1_-_183559693 | 0.38 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr9_+_135037334 | 0.38 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr8_+_21777243 | 0.36 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr21_+_42741979 | 0.36 |
ENST00000543692.1
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr7_+_149411860 | 0.35 |
ENST00000486744.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr8_+_128427857 | 0.35 |
ENST00000391675.1
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr14_-_24615523 | 0.34 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr19_-_3551043 | 0.34 |
ENST00000589995.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr16_+_88704978 | 0.32 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr2_+_231280954 | 0.32 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr19_-_41859814 | 0.32 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr19_+_48898132 | 0.32 |
ENST00000263269.3
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| chr17_+_40440481 | 0.32 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr16_+_81812863 | 0.32 |
ENST00000359376.3
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr8_+_86376081 | 0.31 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr14_-_75078725 | 0.31 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr16_-_66959429 | 0.31 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr2_+_242127924 | 0.30 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr7_+_75911902 | 0.30 |
ENST00000413003.1
|
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr1_-_47082495 | 0.30 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr19_-_38747172 | 0.30 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr19_+_42381173 | 0.30 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr5_+_98264867 | 0.30 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr20_-_4795747 | 0.30 |
ENST00000379376.2
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr15_-_75017711 | 0.29 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr6_-_31138439 | 0.29 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr7_-_151107767 | 0.29 |
ENST00000477459.1
|
WDR86
|
WD repeat domain 86 |
| chr1_-_183560011 | 0.29 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr8_-_145641864 | 0.29 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr19_-_19051927 | 0.28 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila) |
| chr19_-_50400212 | 0.28 |
ENST00000391826.2
|
IL4I1
|
interleukin 4 induced 1 |
| chr16_+_27279526 | 0.28 |
ENST00000566854.1
|
CTD-3203P2.2
|
HCG1815999; Uncharacterized protein |
| chr19_-_55677920 | 0.27 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr5_+_154173697 | 0.27 |
ENST00000518742.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr12_+_120740119 | 0.26 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr20_-_48532019 | 0.26 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr10_+_99609996 | 0.26 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr9_-_117267717 | 0.26 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr5_-_150466692 | 0.26 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_+_74702214 | 0.26 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr19_+_35940486 | 0.25 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr5_+_60240943 | 0.25 |
ENST00000296597.5
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr19_+_35485682 | 0.25 |
ENST00000599564.1
|
GRAMD1A
|
GRAM domain containing 1A |
| chr16_-_4466622 | 0.25 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr4_+_74735102 | 0.24 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr22_+_35796056 | 0.24 |
ENST00000216122.4
|
MCM5
|
minichromosome maintenance complex component 5 |
| chr19_-_8567478 | 0.24 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr22_+_44319648 | 0.24 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr1_+_28879588 | 0.24 |
ENST00000373830.3
|
TRNAU1AP
|
tRNA selenocysteine 1 associated protein 1 |
| chr2_-_70780770 | 0.24 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr12_+_70760056 | 0.23 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr1_+_901847 | 0.23 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr5_-_150467221 | 0.23 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr13_+_53602894 | 0.23 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr7_+_120629653 | 0.23 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr2_-_27718052 | 0.23 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr2_-_224702740 | 0.23 |
ENST00000444408.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr17_-_9479128 | 0.22 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr10_+_13142075 | 0.22 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr16_-_28192360 | 0.22 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr8_-_62559366 | 0.22 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr4_+_2043689 | 0.22 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr15_+_92397051 | 0.22 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr1_-_209824643 | 0.22 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr19_-_33716750 | 0.22 |
ENST00000253188.4
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chr9_+_95909309 | 0.22 |
ENST00000366188.2
|
RP11-370F5.4
|
RP11-370F5.4 |
| chr19_-_3786354 | 0.22 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr10_+_13141585 | 0.22 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr22_+_35796108 | 0.22 |
ENST00000382011.5
ENST00000416905.1 |
MCM5
|
minichromosome maintenance complex component 5 |
| chr14_+_24616588 | 0.22 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr20_+_33462888 | 0.22 |
ENST00000336325.4
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr1_-_47655686 | 0.22 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr3_+_182971335 | 0.21 |
ENST00000464191.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr3_+_182971583 | 0.21 |
ENST00000460419.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr12_+_93115281 | 0.21 |
ENST00000549856.1
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chrX_+_68048803 | 0.21 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr22_-_37640456 | 0.21 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr19_-_3786253 | 0.20 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_123485102 | 0.20 |
ENST00000488323.1
ENST00000223026.4 |
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr9_+_130911770 | 0.20 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr1_+_207494853 | 0.20 |
ENST00000367064.3
ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr2_+_202047596 | 0.20 |
ENST00000286186.6
ENST00000360132.3 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr17_+_12569306 | 0.20 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr9_+_130911723 | 0.20 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr4_-_74904398 | 0.20 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr20_-_48330377 | 0.20 |
ENST00000371711.4
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr5_+_53686658 | 0.19 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr10_+_112257596 | 0.19 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr20_+_49411543 | 0.19 |
ENST00000609336.1
ENST00000445038.1 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr19_+_45147098 | 0.19 |
ENST00000425690.3
ENST00000344956.4 ENST00000403059.4 |
PVR
|
poliovirus receptor |
| chr16_+_28835437 | 0.19 |
ENST00000568266.1
|
ATXN2L
|
ataxin 2-like |
| chr2_+_219081817 | 0.19 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr16_+_57139933 | 0.19 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr16_-_73082274 | 0.19 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr19_-_55677999 | 0.19 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr19_-_56709162 | 0.18 |
ENST00000589938.1
ENST00000587032.2 ENST00000586855.2 |
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr11_+_818902 | 0.18 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chr22_-_37640277 | 0.18 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr10_+_22605304 | 0.18 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr10_+_13142225 | 0.18 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr11_-_104840093 | 0.18 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_61197508 | 0.18 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr19_-_43835582 | 0.18 |
ENST00000595748.1
|
CTC-490G23.2
|
CTC-490G23.2 |
| chr19_-_45927097 | 0.18 |
ENST00000340192.7
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr19_-_19049791 | 0.18 |
ENST00000594439.1
ENST00000221222.11 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr16_+_31724618 | 0.17 |
ENST00000530881.1
ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr16_+_28835766 | 0.17 |
ENST00000564656.1
|
ATXN2L
|
ataxin 2-like |
| chr1_-_33283754 | 0.17 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr12_-_62997214 | 0.17 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr2_+_202047843 | 0.17 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr1_+_24117627 | 0.17 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr8_-_90769422 | 0.17 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr11_+_46639071 | 0.17 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr11_-_61560053 | 0.17 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chr7_-_151107106 | 0.17 |
ENST00000334493.6
|
WDR86
|
WD repeat domain 86 |
| chr19_+_859654 | 0.17 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chr1_-_45965525 | 0.17 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr3_+_52232102 | 0.17 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr1_-_2126192 | 0.17 |
ENST00000378546.4
|
C1orf86
|
chromosome 1 open reading frame 86 |
| chr9_+_130922537 | 0.16 |
ENST00000372994.1
|
C9orf16
|
chromosome 9 open reading frame 16 |
| chr19_-_36505098 | 0.16 |
ENST00000252984.7
ENST00000486389.1 ENST00000378875.3 ENST00000485128.1 |
ALKBH6
|
alkB, alkylation repair homolog 6 (E. coli) |
| chr16_-_4465886 | 0.16 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr15_-_30686052 | 0.16 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr18_-_19283649 | 0.16 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chrX_+_103173457 | 0.16 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr2_-_97534312 | 0.16 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr12_+_7023491 | 0.16 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr1_-_28559502 | 0.16 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr9_+_107509944 | 0.16 |
ENST00000374767.4
|
NIPSNAP3A
|
nipsnap homolog 3A (C. elegans) |
| chr3_+_23847394 | 0.16 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr5_+_170846640 | 0.16 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr17_-_36997708 | 0.16 |
ENST00000398575.4
|
C17orf98
|
chromosome 17 open reading frame 98 |
| chr14_+_32798462 | 0.15 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_+_96533621 | 0.15 |
ENST00000542517.1
ENST00000506569.1 |
EPHA6
|
EPH receptor A6 |
| chr19_-_38746979 | 0.15 |
ENST00000591291.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_-_6545502 | 0.15 |
ENST00000535355.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr10_-_7708918 | 0.15 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr17_+_66255310 | 0.15 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr16_-_1429010 | 0.15 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr5_+_10564432 | 0.15 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr3_+_46618727 | 0.15 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr3_+_182971018 | 0.15 |
ENST00000326505.3
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr1_-_27682962 | 0.15 |
ENST00000486046.1
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr19_+_1103936 | 0.15 |
ENST00000354171.8
ENST00000589115.1 |
GPX4
|
glutathione peroxidase 4 |
| chr19_-_4831701 | 0.15 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr7_+_149412094 | 0.15 |
ENST00000255992.10
ENST00000319551.8 |
KRBA1
|
KRAB-A domain containing 1 |
| chr3_+_49058444 | 0.15 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr16_-_81110683 | 0.14 |
ENST00000565253.1
ENST00000378611.4 ENST00000299578.5 |
C16orf46
|
chromosome 16 open reading frame 46 |
| chr6_+_31950150 | 0.14 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr11_+_61197572 | 0.14 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chrX_-_49041242 | 0.14 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr7_-_5463175 | 0.14 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr2_+_74153953 | 0.14 |
ENST00000264093.4
ENST00000348222.1 |
DGUOK
|
deoxyguanosine kinase |
| chr19_-_46145696 | 0.14 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_+_48824711 | 0.14 |
ENST00000599704.1
|
EMP3
|
epithelial membrane protein 3 |
| chr19_-_1513188 | 0.14 |
ENST00000330475.4
|
ADAMTSL5
|
ADAMTS-like 5 |
| chr8_+_145582217 | 0.14 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr16_-_88923285 | 0.14 |
ENST00000542788.1
ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr1_+_183441500 | 0.14 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr19_+_54371114 | 0.14 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr4_-_74964904 | 0.14 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr19_-_5978144 | 0.14 |
ENST00000340578.6
ENST00000541471.1 ENST00000591736.1 ENST00000587479.1 |
RANBP3
|
RAN binding protein 3 |
| chr15_-_72767490 | 0.14 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr11_+_61560348 | 0.14 |
ENST00000535723.1
ENST00000574708.1 |
FEN1
FADS2
|
flap structure-specific endonuclease 1 fatty acid desaturase 2 |
| chr1_-_155232047 | 0.14 |
ENST00000302631.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr14_+_92789498 | 0.14 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr11_-_46142505 | 0.14 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_79660798 | 0.14 |
ENST00000571237.1
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr11_-_69490135 | 0.13 |
ENST00000542341.1
|
ORAOV1
|
oral cancer overexpressed 1 |
| chr4_-_146859623 | 0.13 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr17_+_46970127 | 0.13 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr19_+_36605850 | 0.13 |
ENST00000221855.3
|
TBCB
|
tubulin folding cofactor B |
| chr7_+_69064566 | 0.13 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr8_+_32406179 | 0.13 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.6 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.7 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.3 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.1 | 0.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.3 | GO:0046732 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.5 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.3 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.3 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.3 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.6 | GO:0006048 | fructose 6-phosphate metabolic process(GO:0006002) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.2 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.6 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0071224 | positive regulation of immature T cell proliferation(GO:0033091) cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:1902563 | regulation of neutrophil activation(GO:1902563) |
| 0.0 | 1.7 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.0 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 1.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.2 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) linoleoyl-CoA desaturase activity(GO:0016213) 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:1904315 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 2.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |