Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
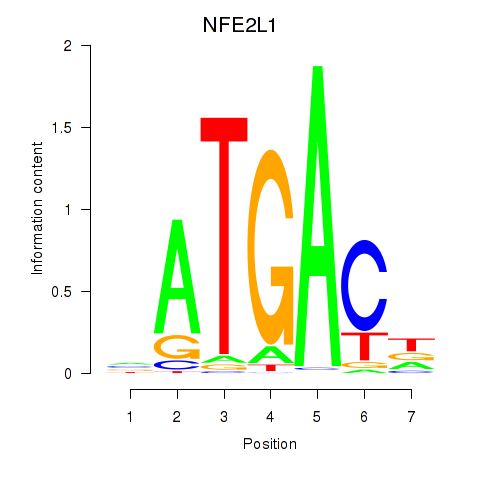
Results for NFE2L1
Z-value: 1.23
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | nuclear factor, erythroid 2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46133307_46133348 | -0.91 | 8.8e-02 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_57308979 | 1.24 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr19_-_36019123 | 0.93 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr17_-_34207295 | 0.88 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr16_+_31044413 | 0.85 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr2_-_158345341 | 0.84 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr10_-_10504285 | 0.69 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr3_-_114790179 | 0.65 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_-_44160768 | 0.60 |
ENST00000593447.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr18_-_2982869 | 0.58 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr15_+_89182178 | 0.56 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 0.54 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr14_-_77542485 | 0.49 |
ENST00000556781.1
ENST00000557526.1 ENST00000555512.1 |
RP11-7F17.3
|
RP11-7F17.3 |
| chr6_+_27114861 | 0.49 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr6_-_32784687 | 0.49 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr16_-_48281305 | 0.48 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr19_+_4402659 | 0.48 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr1_-_100643765 | 0.47 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr2_+_169926047 | 0.45 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_109399682 | 0.45 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr15_+_89181974 | 0.45 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr4_+_185734773 | 0.45 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr16_+_31044812 | 0.43 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr12_-_123187890 | 0.42 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chrX_-_48814278 | 0.42 |
ENST00000455452.1
|
OTUD5
|
OTU domain containing 5 |
| chr15_+_101389945 | 0.41 |
ENST00000561231.1
ENST00000559331.1 ENST00000558254.1 |
RP11-66B24.2
|
RP11-66B24.2 |
| chr22_+_22723969 | 0.40 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr19_-_44160692 | 0.39 |
ENST00000595038.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr7_-_34978980 | 0.39 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr4_+_169013666 | 0.38 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr9_-_100684845 | 0.38 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr3_-_108248169 | 0.38 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr17_-_8021710 | 0.37 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr6_-_43027105 | 0.37 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr3_-_171489085 | 0.36 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr16_+_3068393 | 0.36 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr17_-_71223839 | 0.36 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr10_-_69991865 | 0.36 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr6_+_31916733 | 0.35 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr1_-_204135450 | 0.35 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr2_-_25451065 | 0.34 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr7_-_7782204 | 0.34 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr22_+_30805086 | 0.33 |
ENST00000439838.1
ENST00000439023.3 |
RP4-539M6.19
|
Uncharacterized protein |
| chr9_-_132515302 | 0.33 |
ENST00000340607.4
|
PTGES
|
prostaglandin E synthase |
| chr2_+_87755054 | 0.33 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chrX_+_135279179 | 0.33 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr19_-_48018203 | 0.32 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr19_-_39881669 | 0.32 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr9_-_34665983 | 0.32 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr19_+_8455077 | 0.32 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr6_-_15586238 | 0.31 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr16_+_8814563 | 0.31 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr1_-_31661000 | 0.30 |
ENST00000263693.1
ENST00000398657.2 ENST00000526106.1 |
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr11_+_65154070 | 0.30 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr2_+_87808725 | 0.30 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr7_+_871559 | 0.29 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chrX_-_48814810 | 0.29 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr15_-_81616446 | 0.29 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr6_-_43595039 | 0.29 |
ENST00000307114.7
|
GTPBP2
|
GTP binding protein 2 |
| chr11_+_35222629 | 0.29 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_54813229 | 0.29 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr12_-_53228079 | 0.29 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr2_+_219110149 | 0.29 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr7_-_41742697 | 0.29 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr11_-_119999611 | 0.28 |
ENST00000529044.1
|
TRIM29
|
tripartite motif containing 29 |
| chr17_+_73663470 | 0.28 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr1_+_26605618 | 0.27 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr6_-_31107127 | 0.27 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr19_-_39881777 | 0.27 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr19_+_18699599 | 0.27 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr1_-_198906528 | 0.27 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr2_+_182850743 | 0.27 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr6_+_163837347 | 0.27 |
ENST00000544436.1
|
QKI
|
QKI, KH domain containing, RNA binding |
| chr11_-_66103932 | 0.26 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr16_-_46797149 | 0.26 |
ENST00000536476.1
|
MYLK3
|
myosin light chain kinase 3 |
| chr11_-_119999539 | 0.26 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chr2_+_113735575 | 0.26 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr2_-_158345462 | 0.26 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr8_-_107782463 | 0.26 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr2_+_54198210 | 0.25 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr7_+_150811705 | 0.25 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr12_+_40787194 | 0.25 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr2_+_201994042 | 0.25 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_159750776 | 0.25 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr17_+_73663402 | 0.25 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr17_-_57784755 | 0.25 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr2_-_68547061 | 0.25 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr20_+_60174827 | 0.24 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr22_+_38142235 | 0.24 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr9_+_134065506 | 0.24 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr2_+_169923577 | 0.24 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr19_+_18699535 | 0.24 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr19_+_40477062 | 0.24 |
ENST00000455878.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr12_+_112563303 | 0.24 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr9_+_140083099 | 0.24 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr2_+_169923504 | 0.24 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr17_-_26220366 | 0.24 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr19_+_9361606 | 0.23 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr2_+_182850551 | 0.23 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr8_-_123139423 | 0.23 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr11_+_65337901 | 0.23 |
ENST00000309328.3
ENST00000531405.1 ENST00000527920.1 ENST00000526877.1 ENST00000533115.1 ENST00000526433.1 |
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1 |
| chrX_-_70326455 | 0.23 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr22_-_39097527 | 0.23 |
ENST00000417712.1
|
JOSD1
|
Josephin domain containing 1 |
| chr4_-_69083720 | 0.22 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr11_+_74811578 | 0.22 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr11_+_108093755 | 0.22 |
ENST00000527891.1
ENST00000532931.1 |
ATM
|
ataxia telangiectasia mutated |
| chr14_+_29241910 | 0.22 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr22_+_20861858 | 0.22 |
ENST00000414658.1
ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15
|
mediator complex subunit 15 |
| chr7_-_30008849 | 0.22 |
ENST00000409497.1
|
SCRN1
|
secernin 1 |
| chr10_+_115511434 | 0.22 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr12_-_118628315 | 0.21 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr3_-_110612323 | 0.21 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr10_-_49860525 | 0.21 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr1_-_224624730 | 0.21 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr10_+_97759848 | 0.21 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr22_+_20862321 | 0.21 |
ENST00000541476.1
ENST00000438962.1 |
MED15
|
mediator complex subunit 15 |
| chr12_-_123565834 | 0.21 |
ENST00000546049.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr6_+_31939608 | 0.20 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr9_-_95244781 | 0.20 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr2_-_136594740 | 0.20 |
ENST00000264162.2
|
LCT
|
lactase |
| chr12_+_112563335 | 0.20 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr3_-_112218378 | 0.20 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr6_-_133055815 | 0.20 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr10_+_35464513 | 0.19 |
ENST00000494479.1
ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_13272904 | 0.19 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr11_+_63742050 | 0.19 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr6_-_134639180 | 0.19 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_+_44100632 | 0.19 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr2_+_16080659 | 0.19 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr19_+_42773371 | 0.19 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr19_-_1132207 | 0.19 |
ENST00000438103.2
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr2_+_234602305 | 0.19 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr1_+_159750720 | 0.19 |
ENST00000368109.1
ENST00000368108.3 |
DUSP23
|
dual specificity phosphatase 23 |
| chr22_-_30968813 | 0.19 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_-_113258090 | 0.19 |
ENST00000309276.6
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr19_+_40476912 | 0.18 |
ENST00000157812.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr2_-_99871570 | 0.18 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr6_+_53794780 | 0.18 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr16_+_2286726 | 0.18 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr11_+_58390132 | 0.18 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chr2_+_234160217 | 0.18 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr16_+_15596123 | 0.18 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr4_+_26344754 | 0.18 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_-_70263979 | 0.18 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr21_-_43816052 | 0.18 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr5_-_180242534 | 0.18 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chrX_+_135278908 | 0.18 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr20_+_44563267 | 0.18 |
ENST00000372409.3
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1 |
| chr6_-_32145861 | 0.18 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr17_-_73663245 | 0.18 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr9_+_35538616 | 0.18 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr12_-_52887034 | 0.18 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr7_+_96634850 | 0.18 |
ENST00000518156.2
|
DLX6
|
distal-less homeobox 6 |
| chr12_-_123201337 | 0.18 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr1_-_6445809 | 0.18 |
ENST00000377855.2
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr2_+_87754887 | 0.17 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr5_+_36608422 | 0.17 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_+_28256874 | 0.17 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr2_+_27651519 | 0.17 |
ENST00000379863.3
|
NRBP1
|
nuclear receptor binding protein 1 |
| chr11_-_87908600 | 0.17 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chrX_+_107069063 | 0.17 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr18_+_34124507 | 0.17 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr19_+_55996316 | 0.17 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr17_-_40170506 | 0.16 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr11_+_64052294 | 0.16 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr14_+_21492331 | 0.16 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr1_+_45805728 | 0.16 |
ENST00000539779.1
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr17_+_6916527 | 0.16 |
ENST00000552321.1
|
RNASEK
|
ribonuclease, RNase K |
| chr11_-_62599505 | 0.16 |
ENST00000377897.4
ENST00000394690.1 ENST00000541317.1 ENST00000294179.3 |
STX5
|
syntaxin 5 |
| chr1_-_214638146 | 0.16 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr7_+_80275752 | 0.16 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_-_220119280 | 0.16 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr11_-_128894053 | 0.16 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_+_74701062 | 0.16 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr9_+_6757634 | 0.16 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chrX_+_100224676 | 0.16 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr20_-_1447467 | 0.16 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr10_-_51958906 | 0.16 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr2_-_106015527 | 0.16 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr19_+_50706866 | 0.16 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr20_+_43992094 | 0.16 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chrX_-_128782722 | 0.16 |
ENST00000427399.1
|
APLN
|
apelin |
| chr19_-_49843539 | 0.15 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr19_+_44100727 | 0.15 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr2_+_220071490 | 0.15 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr2_+_87754989 | 0.15 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr9_-_117853297 | 0.15 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr1_+_114522049 | 0.15 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr17_+_33914276 | 0.15 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr1_-_23520755 | 0.15 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr14_+_23938891 | 0.15 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chr16_+_19079215 | 0.15 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr6_-_3157760 | 0.15 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr22_-_30968839 | 0.15 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr2_+_234160340 | 0.15 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr8_+_24151620 | 0.15 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr6_-_31619742 | 0.15 |
ENST00000433828.1
ENST00000456286.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_115608304 | 0.15 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr6_-_31940065 | 0.15 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr12_-_53171128 | 0.15 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr5_+_142286887 | 0.15 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 1.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 0.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 1.0 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.3 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.3 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:0060298 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) sarcomerogenesis(GO:0048769) positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 0.4 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.2 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0042335 | cuticle development(GO:0042335) hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.4 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.4 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.3 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0043132 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) NAD transport(GO:0043132) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.6 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:1901204 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 1.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 1.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.0 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.9 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.3 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.0 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |