Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
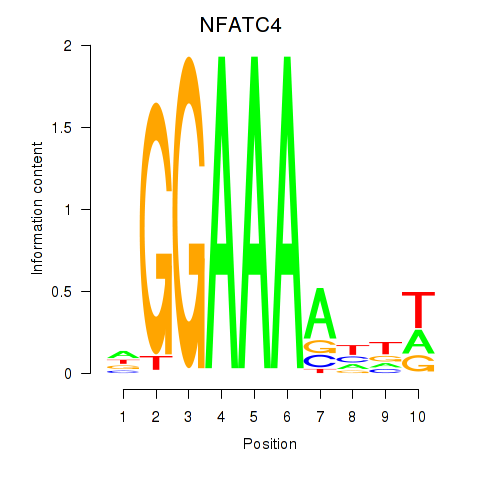
Results for NFATC4
Z-value: 1.38
Transcription factors associated with NFATC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC4
|
ENSG00000100968.9 | nuclear factor of activated T cells 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC4 | hg19_v2_chr14_+_24837226_24837547 | -0.76 | 2.4e-01 | Click! |
Activity profile of NFATC4 motif
Sorted Z-values of NFATC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_89300158 | 1.06 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr6_-_26235206 | 0.93 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr12_-_89918982 | 0.84 |
ENST00000549504.1
|
POC1B
|
POC1 centriolar protein B |
| chr14_+_74551650 | 0.80 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr16_+_53412368 | 0.79 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr15_+_32933866 | 0.78 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr14_+_32546274 | 0.77 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr21_+_17791648 | 0.76 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_29690358 | 0.75 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr17_-_1613663 | 0.73 |
ENST00000330676.6
|
TLCD2
|
TLC domain containing 2 |
| chr11_+_86502085 | 0.67 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr12_-_71551652 | 0.66 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_+_119316721 | 0.64 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr1_+_212782012 | 0.61 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr7_+_23210760 | 0.60 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr6_+_26087646 | 0.59 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr17_+_76311791 | 0.59 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr1_-_145470383 | 0.57 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr8_-_95274536 | 0.55 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr3_-_112329110 | 0.55 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chrX_-_77041685 | 0.55 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr3_-_27764190 | 0.55 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr4_-_71532339 | 0.54 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr10_-_62704005 | 0.54 |
ENST00000337910.5
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr1_+_45274154 | 0.54 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr11_+_77532233 | 0.53 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr9_+_92219919 | 0.53 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr10_-_116444371 | 0.52 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_+_78470530 | 0.52 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr14_+_21156915 | 0.51 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr15_-_37393406 | 0.51 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr5_-_142783175 | 0.51 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr10_-_98347063 | 0.51 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr1_-_150207017 | 0.51 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_+_61869748 | 0.50 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr17_+_44701402 | 0.48 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr20_-_14318248 | 0.47 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr12_+_41086297 | 0.46 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr8_+_98900132 | 0.46 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr11_-_69867159 | 0.45 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chrX_+_100663243 | 0.45 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr1_+_65613513 | 0.45 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr12_-_59314246 | 0.45 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr3_+_118905564 | 0.44 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr14_+_100531615 | 0.44 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr12_-_89918522 | 0.43 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr19_-_33360647 | 0.43 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr14_+_100531738 | 0.42 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr14_-_30396948 | 0.42 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr6_-_26056695 | 0.41 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr17_-_39165366 | 0.40 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr12_-_76461249 | 0.40 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_119316689 | 0.40 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr9_-_98189055 | 0.40 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chr2_+_190541153 | 0.39 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr1_+_24645807 | 0.39 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_219615984 | 0.39 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr11_+_65383227 | 0.39 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr14_+_32546145 | 0.39 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr12_+_41086215 | 0.38 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr7_+_18535893 | 0.37 |
ENST00000432645.2
ENST00000441542.2 |
HDAC9
|
histone deacetylase 9 |
| chr3_-_49837254 | 0.37 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr17_-_5026397 | 0.37 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chr12_-_39734783 | 0.36 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chrX_+_22056165 | 0.36 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_-_153303658 | 0.36 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_+_72112418 | 0.36 |
ENST00000454282.1
|
TNPO1
|
transportin 1 |
| chr16_+_67063036 | 0.36 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr9_-_104145795 | 0.36 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr6_-_34855773 | 0.36 |
ENST00000420584.2
ENST00000361288.4 |
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr1_+_24645865 | 0.35 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr20_-_8000426 | 0.35 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr8_-_25281747 | 0.35 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr1_+_65613217 | 0.35 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr5_+_68462944 | 0.35 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chr14_-_74551096 | 0.35 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr3_-_58200398 | 0.34 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr2_-_190044480 | 0.34 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr19_-_39390440 | 0.34 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr1_-_151299842 | 0.34 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr20_+_57875758 | 0.34 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr1_+_24646002 | 0.34 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr22_-_29107919 | 0.34 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr5_+_145317356 | 0.33 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr3_+_142342228 | 0.32 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr4_-_174254823 | 0.32 |
ENST00000438704.2
|
HMGB2
|
high mobility group box 2 |
| chr6_-_47009996 | 0.32 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr16_-_47493041 | 0.32 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr12_-_68845417 | 0.31 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr5_-_42811986 | 0.31 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr14_+_56078695 | 0.31 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_-_119379427 | 0.31 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr15_+_65914260 | 0.31 |
ENST00000261892.6
ENST00000339868.6 |
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr16_+_75182376 | 0.31 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr11_-_87908600 | 0.31 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.6 |
RAB38
|
RAB38, member RAS oncogene family |
| chr9_-_113342160 | 0.30 |
ENST00000401783.2
ENST00000374461.1 |
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr14_-_77889860 | 0.30 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr11_+_62475130 | 0.30 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr14_-_92413353 | 0.30 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr2_-_191115229 | 0.30 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr14_+_32547434 | 0.30 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_65613340 | 0.30 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr19_-_39390212 | 0.29 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr12_-_90049878 | 0.29 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_49581152 | 0.29 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr19_-_10697895 | 0.29 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr3_-_114035026 | 0.29 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr21_+_17791838 | 0.29 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_+_82767284 | 0.29 |
ENST00000265077.3
|
VCAN
|
versican |
| chr9_-_107754034 | 0.29 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr5_+_43603229 | 0.28 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr14_-_74551172 | 0.28 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr19_+_10959043 | 0.28 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr1_-_63782888 | 0.28 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr1_+_162602244 | 0.28 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr1_+_24646263 | 0.27 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr12_-_90049828 | 0.27 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_-_56492989 | 0.27 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr1_+_24975417 | 0.27 |
ENST00000537199.1
|
SRRM1
|
serine/arginine repetitive matrix 1 |
| chr2_+_191208791 | 0.27 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr5_+_35852797 | 0.26 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr19_-_39390350 | 0.26 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr2_+_223916862 | 0.26 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr17_+_39968926 | 0.26 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr5_+_72112470 | 0.26 |
ENST00000447967.2
ENST00000523768.1 |
TNPO1
|
transportin 1 |
| chr17_-_38938786 | 0.26 |
ENST00000301656.3
|
KRT27
|
keratin 27 |
| chr1_-_161039753 | 0.25 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr20_+_18125727 | 0.25 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr22_+_21319396 | 0.25 |
ENST00000399167.2
ENST00000399163.2 ENST00000441376.2 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr19_+_41856816 | 0.25 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr20_+_35201993 | 0.25 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr1_+_104068562 | 0.25 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr12_-_49582837 | 0.25 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr1_+_104068312 | 0.25 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr5_+_158690089 | 0.24 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr8_-_27468717 | 0.24 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr18_+_52385068 | 0.24 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr19_+_52772821 | 0.24 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr7_-_32529973 | 0.24 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr4_-_141074123 | 0.24 |
ENST00000502696.1
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr3_+_178866199 | 0.24 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr12_-_49582593 | 0.24 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_+_61547894 | 0.24 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr1_+_52521797 | 0.24 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr1_-_221915418 | 0.24 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr12_-_49582978 | 0.24 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr11_-_67141090 | 0.23 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr12_+_27396901 | 0.23 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr6_+_122720681 | 0.23 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr16_+_67381263 | 0.23 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr10_-_123687943 | 0.23 |
ENST00000540606.1
ENST00000455628.1 |
ATE1
|
arginyltransferase 1 |
| chr14_+_75536335 | 0.23 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr11_-_96076334 | 0.22 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr14_+_75536280 | 0.22 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr15_-_77712477 | 0.22 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr10_+_25305524 | 0.22 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr3_-_149051444 | 0.22 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr7_-_92146729 | 0.22 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr1_+_164528866 | 0.22 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr12_-_89919220 | 0.22 |
ENST00000549035.1
ENST00000393179.4 |
POC1B
|
POC1 centriolar protein B |
| chr1_-_161039647 | 0.22 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_-_151148442 | 0.22 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr20_-_17539456 | 0.22 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr1_+_111682058 | 0.22 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr3_+_108321623 | 0.22 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr1_+_24645915 | 0.22 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr14_+_96858433 | 0.21 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chr2_-_207630033 | 0.21 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr19_-_49137790 | 0.21 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_+_13349711 | 0.21 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr9_+_128509624 | 0.21 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr12_+_58335360 | 0.21 |
ENST00000300145.3
|
XRCC6BP1
|
XRCC6 binding protein 1 |
| chr4_+_175204818 | 0.20 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr15_+_59665194 | 0.20 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr11_+_66824303 | 0.20 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr8_-_95449155 | 0.20 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr19_+_16296191 | 0.20 |
ENST00000589852.1
ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32, member A |
| chr3_-_15563229 | 0.20 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr17_+_52978185 | 0.20 |
ENST00000572405.1
ENST00000572158.1 ENST00000540336.1 ENST00000572298.1 ENST00000536554.1 ENST00000575333.1 ENST00000570499.1 ENST00000572576.1 |
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr3_-_50336826 | 0.20 |
ENST00000443842.1
ENST00000354862.4 ENST00000443094.2 ENST00000415204.1 ENST00000336307.1 |
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related) hyaluronoglucosaminidase 3 |
| chr2_-_179343226 | 0.20 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr4_-_77997126 | 0.20 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr12_-_21910775 | 0.20 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr7_-_99527243 | 0.20 |
ENST00000312891.2
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa |
| chr18_+_32556892 | 0.20 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr11_+_65190245 | 0.20 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr11_+_19799327 | 0.20 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr7_-_26904317 | 0.19 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr11_+_77532155 | 0.19 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr6_-_43276535 | 0.19 |
ENST00000372569.3
ENST00000274990.4 |
CRIP3
|
cysteine-rich protein 3 |
| chr11_-_62474803 | 0.19 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr14_+_54863739 | 0.19 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr17_+_54230819 | 0.19 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr17_+_48610074 | 0.19 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr9_-_115818951 | 0.18 |
ENST00000553380.1
ENST00000374227.3 |
ZFP37
|
ZFP37 zinc finger protein |
| chr11_+_114271367 | 0.18 |
ENST00000544582.1
ENST00000545678.1 |
RBM7
|
RNA binding motif protein 7 |
| chr5_-_108063949 | 0.18 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr12_+_9142131 | 0.18 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr15_-_52587945 | 0.18 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr17_+_36584662 | 0.18 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.6 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.2 | 0.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.3 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 1.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.6 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.4 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 0.3 | GO:0071283 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) cellular response to iron(III) ion(GO:0071283) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 1.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.7 | GO:0046874 | quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.2 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.2 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 1.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.5 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.6 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.2 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0008291 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 1.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.4 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.2 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.2 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.4 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 1.1 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.2 | 1.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 1.0 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.5 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.2 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 0.5 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.2 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 2.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |