Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
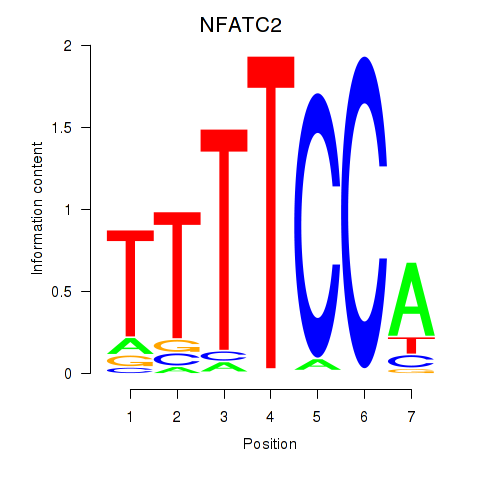
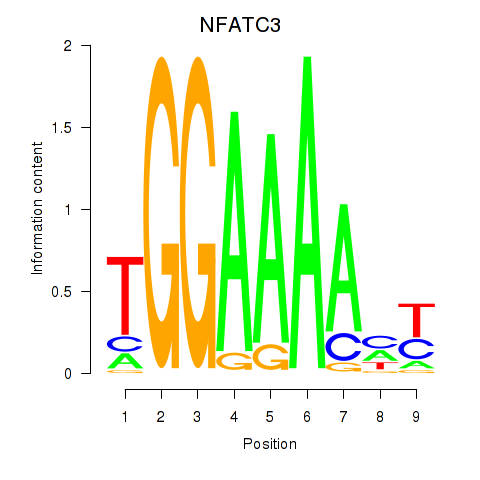
Results for NFATC2_NFATC3
Z-value: 0.18
Transcription factors associated with NFATC2_NFATC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC2
|
ENSG00000101096.15 | nuclear factor of activated T cells 2 |
|
NFATC3
|
ENSG00000072736.14 | nuclear factor of activated T cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC2 | hg19_v2_chr20_-_50159198_50159299 | -0.84 | 1.6e-01 | Click! |
| NFATC3 | hg19_v2_chr16_+_68119764_68119868 | -0.41 | 5.9e-01 | Click! |
Activity profile of NFATC2_NFATC3 motif
Sorted Z-values of NFATC2_NFATC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_139756791 | 0.21 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr15_+_45722727 | 0.20 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr19_-_55881741 | 0.14 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr9_+_19049372 | 0.14 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr17_+_74372662 | 0.12 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr6_+_138188351 | 0.11 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_-_37374876 | 0.11 |
ENST00000405334.1
|
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr5_-_38845812 | 0.10 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr8_-_23712312 | 0.10 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr7_-_41742697 | 0.10 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr14_+_78227105 | 0.10 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr2_-_42160486 | 0.10 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr12_+_113354341 | 0.10 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_156878482 | 0.08 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chrX_-_15683147 | 0.08 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr15_+_63354769 | 0.08 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr1_+_79115503 | 0.08 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr17_+_18647326 | 0.08 |
ENST00000395667.1
ENST00000395665.4 ENST00000308799.4 ENST00000301938.4 |
FBXW10
|
F-box and WD repeat domain containing 10 |
| chr16_+_15528332 | 0.07 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr12_+_75874580 | 0.07 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr15_-_40401062 | 0.06 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr11_-_124806297 | 0.06 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr16_+_56642041 | 0.06 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr17_+_79495397 | 0.06 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr5_+_38846101 | 0.06 |
ENST00000274276.3
|
OSMR
|
oncostatin M receptor |
| chr6_+_31105426 | 0.06 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr9_-_21077939 | 0.06 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr6_+_53794780 | 0.06 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr3_-_156878540 | 0.06 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr3_-_161089289 | 0.06 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr12_-_7244469 | 0.06 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr6_-_13290684 | 0.06 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr11_+_18417948 | 0.06 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr14_-_54955376 | 0.06 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr11_+_18417813 | 0.06 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chr6_+_138188378 | 0.06 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_-_61735029 | 0.05 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr18_-_25739260 | 0.05 |
ENST00000413878.1
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr5_+_49962772 | 0.05 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_-_156675564 | 0.05 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr12_+_75874984 | 0.05 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr12_+_75874460 | 0.05 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr1_+_44401479 | 0.05 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr19_-_39735646 | 0.05 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr16_-_3306587 | 0.05 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr10_-_49860525 | 0.05 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr10_+_3109695 | 0.05 |
ENST00000381125.4
|
PFKP
|
phosphofructokinase, platelet |
| chr2_-_191885686 | 0.05 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chrX_+_133941218 | 0.05 |
ENST00000370784.4
ENST00000370785.3 |
FAM122C
|
family with sequence similarity 122C |
| chr17_+_74261413 | 0.05 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr19_+_18726786 | 0.05 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr17_+_6915730 | 0.04 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr3_+_140981456 | 0.04 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr17_+_7483761 | 0.04 |
ENST00000584180.1
|
CD68
|
CD68 molecule |
| chr7_-_122342988 | 0.04 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr6_+_106534192 | 0.04 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr7_+_120629653 | 0.04 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr11_-_2950642 | 0.04 |
ENST00000314222.4
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2 |
| chr6_+_144665237 | 0.04 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr5_+_135394840 | 0.04 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr13_-_41111323 | 0.04 |
ENST00000595486.1
|
AL133318.1
|
Uncharacterized protein |
| chr14_+_103589789 | 0.04 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr14_+_24837226 | 0.04 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr16_+_57139933 | 0.04 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr1_+_149239529 | 0.04 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr6_-_159065741 | 0.04 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr6_+_31916733 | 0.04 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr1_+_67632083 | 0.04 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr4_-_11431389 | 0.04 |
ENST00000002596.5
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr19_+_7598890 | 0.04 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr5_-_176923803 | 0.04 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr9_-_16705069 | 0.04 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr8_-_73793975 | 0.04 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr8_-_133637624 | 0.04 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr15_+_57891609 | 0.04 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr1_-_161207875 | 0.04 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_+_22766766 | 0.04 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr3_+_14444063 | 0.04 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr12_+_34175398 | 0.04 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr1_-_156675368 | 0.04 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr15_+_63334831 | 0.04 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr6_-_134495992 | 0.04 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_-_158526756 | 0.04 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr6_-_62996066 | 0.04 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr13_-_33924755 | 0.03 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr6_-_112081113 | 0.03 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr10_-_128359074 | 0.03 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr3_-_71632894 | 0.03 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr5_+_155753745 | 0.03 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr12_-_54779511 | 0.03 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr10_-_7453445 | 0.03 |
ENST00000379713.3
ENST00000397167.1 ENST00000397160.3 |
SFMBT2
|
Scm-like with four mbt domains 2 |
| chr18_-_19748331 | 0.03 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chrX_+_48367338 | 0.03 |
ENST00000359882.4
ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN
|
porcupine homolog (Drosophila) |
| chr11_-_61735103 | 0.03 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr5_+_140220769 | 0.03 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr18_+_3451584 | 0.03 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_-_7245018 | 0.03 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr19_+_7599597 | 0.03 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr3_-_100565249 | 0.03 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr4_-_168155730 | 0.03 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr3_+_9944492 | 0.03 |
ENST00000383814.3
ENST00000454190.2 ENST00000454992.1 |
IL17RE
|
interleukin 17 receptor E |
| chr19_+_1041187 | 0.03 |
ENST00000531467.1
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr1_-_155880672 | 0.03 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr12_+_54393880 | 0.03 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr1_-_161207953 | 0.03 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr12_+_6554021 | 0.03 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr3_+_111578131 | 0.03 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_130279178 | 0.03 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr1_-_161207986 | 0.03 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr16_-_1993124 | 0.03 |
ENST00000473663.1
ENST00000399753.2 ENST00000564908.1 |
MSRB1
|
methionine sulfoxide reductase B1 |
| chr12_+_11802753 | 0.03 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr2_-_163099885 | 0.03 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr11_-_123065989 | 0.03 |
ENST00000448775.2
|
CLMP
|
CXADR-like membrane protein |
| chr5_+_158527485 | 0.03 |
ENST00000517335.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr7_+_28452130 | 0.03 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr12_-_57037284 | 0.03 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr19_+_7599128 | 0.03 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr17_+_74261277 | 0.03 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr17_+_4843352 | 0.03 |
ENST00000573404.1
ENST00000576452.1 |
RNF167
|
ring finger protein 167 |
| chr17_-_61045902 | 0.03 |
ENST00000581596.1
|
RP11-180P8.3
|
RP11-180P8.3 |
| chr1_+_901847 | 0.03 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr4_+_75230853 | 0.03 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr10_+_71561630 | 0.03 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr4_+_26322409 | 0.03 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_-_30685563 | 0.03 |
ENST00000401522.3
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr19_+_51226648 | 0.03 |
ENST00000599973.1
|
CLEC11A
|
C-type lectin domain family 11, member A |
| chr11_-_114271139 | 0.03 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr5_+_32531893 | 0.03 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr15_-_90892669 | 0.03 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr11_-_88070896 | 0.03 |
ENST00000529974.1
ENST00000527018.1 |
CTSC
|
cathepsin C |
| chr1_+_209941942 | 0.03 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr7_+_127881325 | 0.03 |
ENST00000308868.4
|
LEP
|
leptin |
| chr3_+_171844762 | 0.03 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr8_+_92114873 | 0.03 |
ENST00000343709.3
ENST00000448384.2 |
LRRC69
|
leucine rich repeat containing 69 |
| chr17_+_9745786 | 0.03 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr20_+_33464238 | 0.03 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr2_+_120517717 | 0.03 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr17_-_15469590 | 0.03 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr11_-_76381029 | 0.03 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr2_+_201997492 | 0.03 |
ENST00000494258.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chrX_+_102841064 | 0.03 |
ENST00000469586.1
|
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr3_-_122102065 | 0.03 |
ENST00000479899.1
ENST00000291458.5 ENST00000497726.1 |
CCDC58
|
coiled-coil domain containing 58 |
| chr5_+_140213815 | 0.03 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr10_+_105036909 | 0.03 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr12_-_21487829 | 0.03 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr11_-_46113756 | 0.03 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr20_+_57209828 | 0.03 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr12_-_104443890 | 0.03 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr11_-_95523500 | 0.03 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr5_-_176923846 | 0.03 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr10_-_52008313 | 0.03 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr16_-_28192360 | 0.03 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr9_+_112542572 | 0.02 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr4_-_71532601 | 0.02 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_-_153521597 | 0.02 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr16_-_73093597 | 0.02 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr19_+_7599792 | 0.02 |
ENST00000600942.1
ENST00000593924.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr11_-_46142948 | 0.02 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr10_+_47746929 | 0.02 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr14_-_102976135 | 0.02 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr4_+_155665123 | 0.02 |
ENST00000336356.3
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr12_-_30887948 | 0.02 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr12_+_38710555 | 0.02 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr18_-_19748379 | 0.02 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr5_+_38845960 | 0.02 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr14_-_24615523 | 0.02 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr18_+_3247413 | 0.02 |
ENST00000579226.1
ENST00000217652.3 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_133055815 | 0.02 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr12_+_51318513 | 0.02 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr1_-_153521714 | 0.02 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr2_+_162272605 | 0.02 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr6_-_134639042 | 0.02 |
ENST00000461976.2
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chrX_-_10544942 | 0.02 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr15_-_30686052 | 0.02 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr15_+_67358163 | 0.02 |
ENST00000327367.4
|
SMAD3
|
SMAD family member 3 |
| chr1_+_150522222 | 0.02 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr5_-_146461027 | 0.02 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_147162263 | 0.02 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr10_+_114710516 | 0.02 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_127963343 | 0.02 |
ENST00000335247.7
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr2_-_145188137 | 0.02 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_122240792 | 0.02 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr6_+_17393888 | 0.02 |
ENST00000493172.1
ENST00000465994.1 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr6_-_26124138 | 0.02 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr1_+_173991633 | 0.02 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr7_-_105319536 | 0.02 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr5_-_94620239 | 0.02 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_+_42925270 | 0.02 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr5_-_77844974 | 0.02 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr1_-_161208013 | 0.02 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_152879985 | 0.02 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chrX_+_33744503 | 0.02 |
ENST00000439992.1
|
RP11-305F18.1
|
RP11-305F18.1 |
| chr2_-_163100045 | 0.02 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr4_+_26322185 | 0.02 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_+_164528437 | 0.02 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_-_68698222 | 0.02 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr1_+_186265399 | 0.02 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr12_+_112563303 | 0.02 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr2_+_232316906 | 0.02 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr16_-_1993260 | 0.02 |
ENST00000361871.3
|
MSRB1
|
methionine sulfoxide reductase B1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC2_NFATC3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.0 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.0 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.0 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |