Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
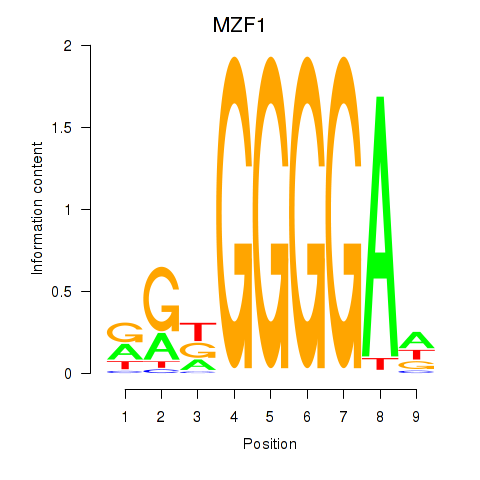
Results for MZF1
Z-value: 0.65
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.4 | myeloid zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg19_v2_chr19_-_59084647_59084721 | 0.89 | 1.1e-01 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_4488872 | 0.57 |
ENST00000237837.1
|
FGF23
|
fibroblast growth factor 23 |
| chr17_-_56591978 | 0.50 |
ENST00000583656.1
|
MTMR4
|
myotubularin related protein 4 |
| chr2_-_182545603 | 0.48 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr13_-_52027134 | 0.44 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr14_-_50999373 | 0.38 |
ENST00000554273.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_+_2264983 | 0.37 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr13_-_52026730 | 0.32 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr5_-_81046904 | 0.31 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr10_+_114710516 | 0.31 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_57030096 | 0.30 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr17_-_46035187 | 0.27 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr1_+_6845578 | 0.27 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_-_61765732 | 0.25 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr18_+_46065570 | 0.24 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chrX_-_45710920 | 0.24 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr9_-_140115775 | 0.23 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr5_-_81046922 | 0.22 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr1_+_6845384 | 0.21 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr5_+_49962495 | 0.20 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_+_88324870 | 0.20 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr2_+_46926326 | 0.20 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr6_-_10412600 | 0.20 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr19_+_50922187 | 0.19 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chrX_+_134654540 | 0.19 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr19_-_4065730 | 0.19 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr10_+_114709999 | 0.19 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr17_-_49337392 | 0.18 |
ENST00000376381.2
ENST00000586178.1 |
MBTD1
|
mbt domain containing 1 |
| chr1_+_6845497 | 0.18 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_-_61765315 | 0.18 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr8_+_145321517 | 0.18 |
ENST00000340210.1
|
SCXB
|
scleraxis homolog B (mouse) |
| chr10_+_102672712 | 0.18 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr14_+_74815116 | 0.18 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr17_+_76311791 | 0.17 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr4_-_56412713 | 0.17 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr17_-_48943706 | 0.17 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr6_-_86353510 | 0.16 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr16_+_30907927 | 0.16 |
ENST00000279804.2
ENST00000395019.3 |
CTF1
|
cardiotrophin 1 |
| chr19_-_42758040 | 0.16 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr16_+_27279526 | 0.16 |
ENST00000566854.1
|
CTD-3203P2.2
|
HCG1815999; Uncharacterized protein |
| chr16_+_30969055 | 0.16 |
ENST00000452917.1
|
SETD1A
|
SET domain containing 1A |
| chr6_-_32160622 | 0.15 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr2_+_5832799 | 0.15 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chrX_+_102841064 | 0.15 |
ENST00000469586.1
|
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chrX_+_107334895 | 0.15 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr2_-_48132924 | 0.15 |
ENST00000403359.3
|
FBXO11
|
F-box protein 11 |
| chr7_-_79082867 | 0.15 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr19_-_36523529 | 0.14 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr7_-_94953878 | 0.14 |
ENST00000222381.3
|
PON1
|
paraoxonase 1 |
| chr7_-_100881041 | 0.14 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr4_-_140222358 | 0.14 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr14_-_50999307 | 0.14 |
ENST00000013125.4
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_+_110790867 | 0.14 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr17_+_29421987 | 0.14 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr20_+_34742650 | 0.14 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chrX_-_131352152 | 0.13 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr1_-_53018654 | 0.13 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr16_-_18937480 | 0.13 |
ENST00000532700.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr15_-_37392724 | 0.13 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr6_-_29600559 | 0.13 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr3_-_88108212 | 0.13 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr6_+_87865262 | 0.13 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr2_+_219283815 | 0.13 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr10_+_114710211 | 0.13 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_+_170075436 | 0.13 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr17_+_38278530 | 0.13 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr17_-_63557309 | 0.13 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr4_-_165305086 | 0.13 |
ENST00000507270.1
ENST00000514618.1 ENST00000503008.1 |
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr9_+_17134980 | 0.12 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr2_+_206950095 | 0.12 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr8_-_38126635 | 0.12 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr17_+_54671047 | 0.12 |
ENST00000332822.4
|
NOG
|
noggin |
| chr17_+_45727204 | 0.12 |
ENST00000290158.4
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr11_+_82868185 | 0.12 |
ENST00000530304.1
ENST00000533018.1 |
PCF11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr10_+_119301928 | 0.12 |
ENST00000553456.3
|
EMX2
|
empty spiracles homeobox 2 |
| chr7_+_77167376 | 0.12 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr17_+_45771420 | 0.12 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr9_+_4985016 | 0.12 |
ENST00000539801.1
|
JAK2
|
Janus kinase 2 |
| chr6_-_62996066 | 0.12 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr9_+_17135016 | 0.12 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr14_+_96968802 | 0.12 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr5_-_180287663 | 0.12 |
ENST00000509066.1
ENST00000504225.1 |
ZFP62
|
ZFP62 zinc finger protein |
| chr2_+_45168875 | 0.12 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr14_+_90864504 | 0.12 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_-_155532484 | 0.12 |
ENST00000368346.3
ENST00000548830.1 |
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr15_-_71055769 | 0.12 |
ENST00000539319.1
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr16_+_77246337 | 0.12 |
ENST00000563157.1
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr17_-_46703826 | 0.11 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr15_+_75487984 | 0.11 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr15_-_75743991 | 0.11 |
ENST00000567289.1
|
SIN3A
|
SIN3 transcription regulator family member A |
| chrX_-_128657457 | 0.11 |
ENST00000371121.3
ENST00000371123.1 ENST00000371122.4 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chrX_-_119695279 | 0.11 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr2_-_61389168 | 0.11 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr1_+_93913665 | 0.11 |
ENST00000271234.7
ENST00000370256.4 ENST00000260506.8 |
FNBP1L
|
formin binding protein 1-like |
| chr16_+_21610797 | 0.11 |
ENST00000358154.3
|
METTL9
|
methyltransferase like 9 |
| chr7_+_65338312 | 0.11 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr2_-_48132814 | 0.11 |
ENST00000316377.4
ENST00000378314.3 |
FBXO11
|
F-box protein 11 |
| chr1_-_11115877 | 0.11 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr2_+_145780739 | 0.11 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_-_141060389 | 0.11 |
ENST00000504448.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr16_+_87425381 | 0.11 |
ENST00000268607.5
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta |
| chr14_+_64971438 | 0.11 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chrX_-_151619746 | 0.11 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr12_-_123011476 | 0.11 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr1_+_155829341 | 0.11 |
ENST00000539162.1
|
SYT11
|
synaptotagmin XI |
| chr10_-_12084770 | 0.11 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr12_+_22778291 | 0.11 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr6_+_30614779 | 0.11 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr10_+_97803151 | 0.11 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr12_-_72057638 | 0.10 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr8_+_136470270 | 0.10 |
ENST00000524199.1
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr19_+_3708338 | 0.10 |
ENST00000590545.1
|
TJP3
|
tight junction protein 3 |
| chr17_-_7166500 | 0.10 |
ENST00000575313.1
ENST00000397317.4 |
CLDN7
|
claudin 7 |
| chr3_-_24536253 | 0.10 |
ENST00000428492.1
ENST00000396671.2 ENST00000431815.1 ENST00000418247.1 ENST00000416420.1 ENST00000356447.4 |
THRB
|
thyroid hormone receptor, beta |
| chr1_-_150208498 | 0.10 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_49365501 | 0.10 |
ENST00000403957.1
ENST00000301061.4 |
WNT10B
|
wingless-type MMTV integration site family, member 10B |
| chr12_+_10366223 | 0.10 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr14_+_96968707 | 0.10 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr7_+_6522922 | 0.10 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr3_-_69171708 | 0.10 |
ENST00000420581.2
|
LMOD3
|
leiomodin 3 (fetal) |
| chr17_-_46667594 | 0.10 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr7_-_76955563 | 0.10 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr19_+_41770236 | 0.10 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_+_76356516 | 0.10 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr22_+_40440804 | 0.10 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr12_+_57916584 | 0.10 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_+_83080724 | 0.10 |
ENST00000548305.1
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr10_-_126849626 | 0.10 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr4_+_153457404 | 0.10 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr12_+_46123448 | 0.10 |
ENST00000334344.6
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr14_-_71107921 | 0.10 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr4_-_100815525 | 0.09 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr16_-_30394143 | 0.09 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr15_-_43212836 | 0.09 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr1_-_226065330 | 0.09 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr19_-_14201507 | 0.09 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr15_-_49103235 | 0.09 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr18_-_51750948 | 0.09 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chrX_-_119694538 | 0.09 |
ENST00000371322.5
|
CUL4B
|
cullin 4B |
| chr7_-_127032363 | 0.09 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr3_+_196466710 | 0.09 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr14_-_65569186 | 0.09 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr1_+_150898733 | 0.09 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chr22_-_41842781 | 0.09 |
ENST00000434408.1
|
TOB2
|
transducer of ERBB2, 2 |
| chr3_+_119814070 | 0.09 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr16_-_1843694 | 0.09 |
ENST00000569769.1
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr8_-_71581377 | 0.09 |
ENST00000276590.4
ENST00000522447.1 |
LACTB2
|
lactamase, beta 2 |
| chr18_+_7946839 | 0.09 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chrX_+_119495934 | 0.09 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr3_-_161089289 | 0.09 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr1_+_109234907 | 0.09 |
ENST00000370025.4
ENST00000370022.5 ENST00000370021.1 |
PRPF38B
|
pre-mRNA processing factor 38B |
| chr8_-_119634141 | 0.09 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr8_-_145018905 | 0.09 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr5_-_180288248 | 0.09 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr1_+_203765437 | 0.09 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chrX_-_47004437 | 0.09 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr16_+_21610879 | 0.09 |
ENST00000396014.4
|
METTL9
|
methyltransferase like 9 |
| chr19_-_6670128 | 0.09 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chrX_-_38186775 | 0.09 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr11_-_64410787 | 0.09 |
ENST00000301894.2
|
NRXN2
|
neurexin 2 |
| chr17_-_56065484 | 0.08 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr5_+_52776449 | 0.08 |
ENST00000396947.3
|
FST
|
follistatin |
| chr11_+_64794991 | 0.08 |
ENST00000352068.5
ENST00000525648.1 |
SNX15
|
sorting nexin 15 |
| chr1_+_113217309 | 0.08 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chrX_-_19140677 | 0.08 |
ENST00000357544.3
ENST00000379869.3 ENST00000360279.4 ENST00000379873.2 ENST00000379878.3 ENST00000354791.3 ENST00000379876.1 |
GPR64
|
G protein-coupled receptor 64 |
| chr8_+_29952914 | 0.08 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr2_-_197458323 | 0.08 |
ENST00000452031.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr10_-_99094458 | 0.08 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr12_-_15114603 | 0.08 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr7_+_8008418 | 0.08 |
ENST00000223145.5
|
GLCCI1
|
glucocorticoid induced transcript 1 |
| chr1_-_200379129 | 0.08 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr3_+_110790715 | 0.08 |
ENST00000319792.3
|
PVRL3
|
poliovirus receptor-related 3 |
| chr7_-_127032741 | 0.08 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr1_+_97188188 | 0.08 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr11_-_8832521 | 0.08 |
ENST00000530438.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr14_+_31494841 | 0.08 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr17_+_38278826 | 0.08 |
ENST00000577454.1
ENST00000578648.1 ENST00000579565.1 |
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr15_+_75640068 | 0.08 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr11_+_63448918 | 0.08 |
ENST00000341307.2
ENST00000356000.3 ENST00000542238.1 |
RTN3
|
reticulon 3 |
| chr17_+_42634844 | 0.08 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr1_+_66999268 | 0.08 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr7_+_128312346 | 0.08 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr1_-_150208412 | 0.08 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr14_+_75745477 | 0.08 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr12_-_123011536 | 0.08 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr17_+_29421900 | 0.08 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr1_-_200379180 | 0.08 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr11_+_63448955 | 0.08 |
ENST00000377819.5
ENST00000339997.4 ENST00000540798.1 ENST00000545432.1 ENST00000543552.1 ENST00000537981.1 |
RTN3
|
reticulon 3 |
| chr11_+_82868030 | 0.08 |
ENST00000298281.4
ENST00000530660.1 |
PCF11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr3_-_93692781 | 0.08 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr8_-_117886563 | 0.08 |
ENST00000519837.1
ENST00000522699.1 |
RAD21
|
RAD21 homolog (S. pombe) |
| chr2_+_159313452 | 0.08 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr17_+_34901353 | 0.08 |
ENST00000593016.1
|
GGNBP2
|
gametogenetin binding protein 2 |
| chr2_-_230786378 | 0.07 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr14_+_23790655 | 0.07 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr3_-_182698381 | 0.07 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr13_-_31039375 | 0.07 |
ENST00000399494.1
|
HMGB1
|
high mobility group box 1 |
| chr14_+_24099318 | 0.07 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr6_+_32936942 | 0.07 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr19_-_15442701 | 0.07 |
ENST00000594841.1
ENST00000601941.1 |
BRD4
|
bromodomain containing 4 |
| chr7_+_26241310 | 0.07 |
ENST00000396386.2
|
CBX3
|
chromobox homolog 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.7 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.0 | GO:0046543 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.3 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0043449 | insecticide metabolic process(GO:0017143) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:1902728 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.2 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:0044727 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0051885 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of anagen(GO:0051885) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.0 | GO:0061010 | external genitalia morphogenesis(GO:0035261) gall bladder development(GO:0061010) |
| 0.0 | 0.0 | GO:0061564 | axon development(GO:0061564) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.0 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:2001076 | thorax and anterior abdomen determination(GO:0007356) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0051694 | positive regulation of skeletal muscle fiber development(GO:0048743) pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:1903440 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |