Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
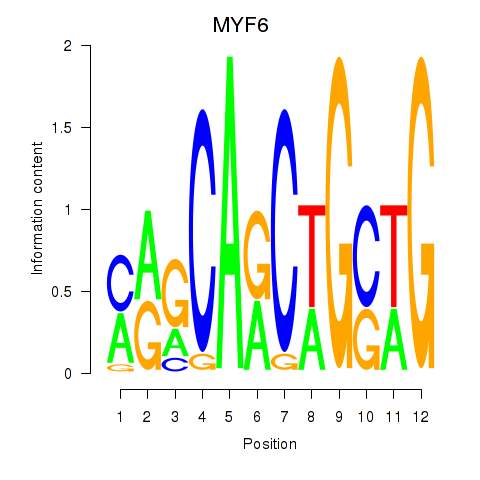
Results for MYF6
Z-value: 1.27
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | myogenic factor 6 |
Activity profile of MYF6 motif
Sorted Z-values of MYF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_7005785 | 1.32 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr8_-_23712312 | 1.27 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr17_+_68165657 | 1.26 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr5_-_139422654 | 0.97 |
ENST00000289409.4
ENST00000358522.3 ENST00000378238.4 ENST00000289422.7 ENST00000361474.1 ENST00000545385.1 ENST00000394770.1 ENST00000541337.1 |
NRG2
|
neuregulin 2 |
| chr4_+_89299994 | 0.97 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr4_+_89299885 | 0.94 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr8_-_145086922 | 0.89 |
ENST00000530478.1
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr4_+_7194247 | 0.79 |
ENST00000507866.2
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr11_-_627143 | 0.73 |
ENST00000176195.3
|
SCT
|
secretin |
| chr9_-_131940526 | 0.69 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr10_-_79397547 | 0.68 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr15_+_90744745 | 0.65 |
ENST00000558051.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr17_+_38171614 | 0.65 |
ENST00000583218.1
ENST00000394149.3 |
CSF3
|
colony stimulating factor 3 (granulocyte) |
| chr20_-_60942326 | 0.62 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr2_-_231084820 | 0.56 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr11_-_33913708 | 0.56 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr10_-_90967063 | 0.55 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr2_-_231084659 | 0.54 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr20_-_23669590 | 0.53 |
ENST00000217423.3
|
CST4
|
cystatin S |
| chr10_-_79397740 | 0.51 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr22_-_50964849 | 0.50 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr12_-_76425368 | 0.50 |
ENST00000602540.1
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1 |
| chr17_-_47308128 | 0.50 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr20_-_60942361 | 0.49 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr19_+_51153045 | 0.49 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr22_-_21905120 | 0.48 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr19_-_18391708 | 0.48 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr4_-_168155730 | 0.48 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_+_32006159 | 0.46 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chrX_-_109590174 | 0.45 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr19_+_35521616 | 0.44 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr11_-_33891362 | 0.44 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr17_+_74261413 | 0.44 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr4_-_141075330 | 0.44 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr15_+_90744533 | 0.44 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr19_-_44174330 | 0.43 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr6_-_31324943 | 0.43 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr17_+_74381343 | 0.42 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr9_-_38069208 | 0.41 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr6_+_32006042 | 0.41 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr22_-_50964558 | 0.41 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_+_35521572 | 0.41 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr17_-_39183452 | 0.40 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr4_-_185395191 | 0.40 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr2_+_206547215 | 0.38 |
ENST00000360409.3
ENST00000540178.1 ENST00000540841.1 ENST00000355117.4 ENST00000450507.1 ENST00000417189.1 |
NRP2
|
neuropilin 2 |
| chr1_-_21503337 | 0.38 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr22_+_31489344 | 0.38 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr4_-_140223614 | 0.38 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_-_231084617 | 0.38 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr19_+_35521699 | 0.37 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr1_+_23037323 | 0.37 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr19_-_44174305 | 0.37 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr2_+_47168313 | 0.37 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr15_+_41221536 | 0.37 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr9_-_128003606 | 0.36 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr18_+_61637159 | 0.35 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr8_-_131028782 | 0.35 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr7_+_100210133 | 0.34 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr2_+_79740118 | 0.34 |
ENST00000496558.1
ENST00000451966.1 |
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr4_+_2043689 | 0.33 |
ENST00000382878.3
ENST00000409248.4 |
C4orf48
|
chromosome 4 open reading frame 48 |
| chr5_+_49962772 | 0.33 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_+_39687596 | 0.33 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr1_-_47082495 | 0.33 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr13_-_44361025 | 0.32 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr2_-_61765732 | 0.32 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr5_-_141704566 | 0.32 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr1_-_144866711 | 0.32 |
ENST00000530130.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr17_+_74261277 | 0.32 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr11_-_10830463 | 0.32 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chrX_-_19905703 | 0.31 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr1_-_154842741 | 0.31 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr20_+_2795609 | 0.31 |
ENST00000554164.1
ENST00000380593.4 |
TMEM239
TMEM239
|
transmembrane protein 239 CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
| chr19_+_1286097 | 0.30 |
ENST00000215368.2
|
EFNA2
|
ephrin-A2 |
| chr20_+_4129426 | 0.30 |
ENST00000339123.6
ENST00000305958.4 ENST00000278795.3 |
SMOX
|
spermine oxidase |
| chrX_-_80457385 | 0.30 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr19_-_40730820 | 0.29 |
ENST00000513948.1
|
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr5_-_94620239 | 0.29 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr19_-_3557570 | 0.29 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr11_+_74951948 | 0.29 |
ENST00000562197.2
|
TPBGL
|
trophoblast glycoprotein-like |
| chr20_+_4129496 | 0.29 |
ENST00000346595.2
|
SMOX
|
spermine oxidase |
| chr3_+_3841108 | 0.29 |
ENST00000319331.3
|
LRRN1
|
leucine rich repeat neuronal 1 |
| chr11_-_46142505 | 0.28 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_7348658 | 0.28 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr1_+_200842083 | 0.28 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr18_+_21269404 | 0.28 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chrX_-_92928557 | 0.28 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr13_-_108518986 | 0.28 |
ENST00000375915.2
|
FAM155A
|
family with sequence similarity 155, member A |
| chr9_+_137533615 | 0.27 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr6_+_126112001 | 0.27 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_+_41103063 | 0.27 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr6_+_42896865 | 0.27 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chrX_+_73641286 | 0.27 |
ENST00000587091.1
|
SLC16A2
|
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr11_-_46142948 | 0.26 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr19_-_12845550 | 0.26 |
ENST00000242784.4
|
C19orf43
|
chromosome 19 open reading frame 43 |
| chr6_-_139695757 | 0.26 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr15_-_43212996 | 0.26 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr11_-_65325430 | 0.26 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr2_+_202937972 | 0.26 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr22_+_18593446 | 0.25 |
ENST00000316027.6
|
TUBA8
|
tubulin, alpha 8 |
| chr16_+_56623433 | 0.25 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr22_+_18593507 | 0.25 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr4_-_140223670 | 0.25 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr6_-_29600559 | 0.25 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr6_+_1312675 | 0.25 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chr19_-_38747172 | 0.25 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr22_+_19705928 | 0.24 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr20_-_23731569 | 0.24 |
ENST00000304749.2
|
CST1
|
cystatin SN |
| chr2_-_178129551 | 0.24 |
ENST00000430047.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr19_-_6720686 | 0.24 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr11_+_62556596 | 0.24 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chrX_+_118108571 | 0.24 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr1_+_43613566 | 0.24 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr13_-_36050819 | 0.24 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr11_-_10829851 | 0.23 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr17_+_7758374 | 0.23 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr2_+_159651821 | 0.23 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr1_-_57045228 | 0.23 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr19_-_19051103 | 0.23 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr19_-_54604083 | 0.23 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr3_-_124774802 | 0.23 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr7_+_128864848 | 0.23 |
ENST00000325006.3
ENST00000446544.2 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr12_-_71003568 | 0.23 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr6_+_33257346 | 0.23 |
ENST00000374606.5
ENST00000374610.2 ENST00000374607.1 |
PFDN6
|
prefoldin subunit 6 |
| chr1_+_173991633 | 0.23 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr17_+_48133459 | 0.22 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr12_-_54813229 | 0.22 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr17_+_79859985 | 0.22 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr14_+_29236269 | 0.22 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr17_-_47308100 | 0.22 |
ENST00000503902.1
ENST00000512250.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr1_+_110754094 | 0.22 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr21_-_16125773 | 0.22 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr2_+_148778570 | 0.22 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr17_-_41132088 | 0.22 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr17_-_42452063 | 0.21 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr11_-_65325664 | 0.21 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr6_+_31105426 | 0.21 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr4_-_46391367 | 0.21 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr4_-_129209221 | 0.21 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr4_+_2043777 | 0.21 |
ENST00000409860.1
|
C4orf48
|
chromosome 4 open reading frame 48 |
| chrX_+_24483338 | 0.21 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr19_-_12845467 | 0.21 |
ENST00000592273.1
ENST00000588213.1 |
C19orf43
|
chromosome 19 open reading frame 43 |
| chr15_+_24803299 | 0.21 |
ENST00000568609.1
ENST00000569908.1 ENST00000567647.1 ENST00000565241.1 |
PWRN1
|
Prader-Willi region non-protein coding RNA 1 |
| chr11_-_115375107 | 0.21 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr19_-_11616645 | 0.21 |
ENST00000293771.5
|
ZNF653
|
zinc finger protein 653 |
| chr2_-_220408430 | 0.21 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr19_-_51014588 | 0.21 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr14_+_103394963 | 0.21 |
ENST00000559525.1
ENST00000559789.1 |
AMN
|
amnion associated transmembrane protein |
| chr5_-_147162078 | 0.20 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr10_+_102758105 | 0.20 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr19_-_3557401 | 0.20 |
ENST00000389395.3
ENST00000398558.4 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr7_+_100860949 | 0.20 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr16_+_230435 | 0.20 |
ENST00000199708.2
|
HBQ1
|
hemoglobin, theta 1 |
| chr1_+_20915409 | 0.20 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chrX_+_118108601 | 0.20 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr19_-_48016023 | 0.20 |
ENST00000598615.1
ENST00000597118.1 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr18_-_25739260 | 0.20 |
ENST00000413878.1
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr11_-_46142615 | 0.20 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr11_+_308143 | 0.20 |
ENST00000399817.4
|
IFITM2
|
interferon induced transmembrane protein 2 |
| chr2_+_58655461 | 0.19 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr22_+_38071615 | 0.19 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chrX_-_47489244 | 0.19 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr17_-_7760457 | 0.19 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr17_+_48133330 | 0.19 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr1_-_23340400 | 0.19 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr22_+_30792846 | 0.19 |
ENST00000312932.9
ENST00000428195.1 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr20_-_52790512 | 0.19 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr7_+_96635277 | 0.18 |
ENST00000007660.5
|
DLX6
|
distal-less homeobox 6 |
| chr11_+_71276609 | 0.18 |
ENST00000398531.1
ENST00000376536.4 |
KRTAP5-10
|
keratin associated protein 5-10 |
| chr19_-_17445613 | 0.18 |
ENST00000159087.4
|
ANO8
|
anoctamin 8 |
| chrX_-_153151586 | 0.18 |
ENST00000370060.1
ENST00000370055.1 ENST00000420165.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr12_-_68845165 | 0.18 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr4_-_168155577 | 0.18 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr14_-_99737565 | 0.18 |
ENST00000357195.3
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr19_+_54371114 | 0.18 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr22_-_31328881 | 0.18 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr15_-_43212836 | 0.18 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr19_+_54385439 | 0.18 |
ENST00000536044.1
ENST00000540413.1 ENST00000263431.3 ENST00000419486.1 |
PRKCG
|
protein kinase C, gamma |
| chr17_+_42429493 | 0.18 |
ENST00000586242.1
|
GRN
|
granulin |
| chr19_+_45349630 | 0.18 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr11_-_88070896 | 0.18 |
ENST00000529974.1
ENST00000527018.1 |
CTSC
|
cathepsin C |
| chr2_+_7005959 | 0.18 |
ENST00000442639.1
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr11_-_67188642 | 0.17 |
ENST00000546202.1
ENST00000542876.1 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr9_-_140064496 | 0.17 |
ENST00000371542.3
|
LRRC26
|
leucine rich repeat containing 26 |
| chr1_-_11918988 | 0.17 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr19_+_35940486 | 0.17 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr11_-_47546250 | 0.17 |
ENST00000543178.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr17_+_4802713 | 0.17 |
ENST00000521575.1
ENST00000381365.3 |
C17orf107
|
chromosome 17 open reading frame 107 |
| chr5_+_135385202 | 0.17 |
ENST00000514554.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr5_-_147162263 | 0.17 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr17_-_7760779 | 0.17 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr6_-_31514516 | 0.17 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr18_+_21269556 | 0.17 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr3_+_32280159 | 0.17 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr19_-_51014460 | 0.17 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr1_+_180123969 | 0.17 |
ENST00000367602.3
ENST00000367600.5 |
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chrX_+_48681768 | 0.17 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr19_-_45908292 | 0.17 |
ENST00000360957.5
ENST00000592134.1 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr10_+_12237924 | 0.17 |
ENST00000429258.2
ENST00000281141.4 |
CDC123
|
cell division cycle 123 |
| chr17_-_26903900 | 0.16 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr14_+_52327350 | 0.16 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYF6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.3 | 1.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 0.7 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 | 0.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 0.5 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 1.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 1.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.4 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 0.3 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.8 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.6 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.1 | 0.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 1.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.3 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.1 | 0.2 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 1.0 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.3 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.1 | 0.7 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.3 | GO:0061428 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.1 | 0.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.3 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0030885 | tolerance induction dependent upon immune response(GO:0002461) regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) response to ozone(GO:0010193) operant conditioning(GO:0035106) sperm ejaculation(GO:0042713) positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.4 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.2 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of sulfur amino acid metabolic process(GO:0031337) regulation of homocysteine metabolic process(GO:0050666) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.1 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.9 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0046778 | modulation by virus of host gene expression(GO:0039656) modification by virus of host mRNA processing(GO:0046778) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0032445 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.0 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 1.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 1.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0051459 | regulation of corticotropin secretion(GO:0051459) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:1903764 | hard palate development(GO:0060022) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0072221 | forebrain ventricular zone progenitor cell division(GO:0021869) ascending thin limb development(GO:0072021) distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric ascending thin limb development(GO:0072218) metanephric distal convoluted tubule development(GO:0072221) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 1.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.2 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.1 | 0.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 2.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0098553 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 1.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.6 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.2 | 1.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.8 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.9 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.2 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.0 | GO:0016898 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |