Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
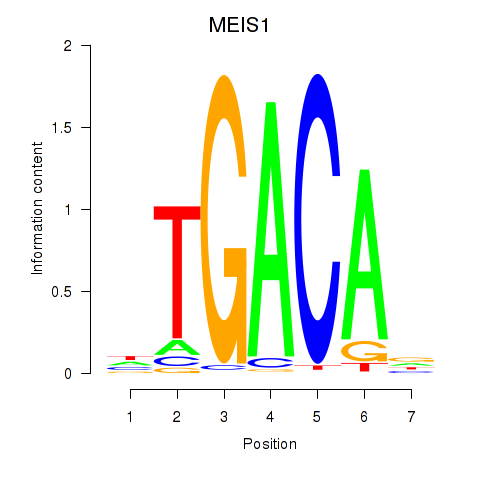
Results for MEIS1
Z-value: 1.22
Transcription factors associated with MEIS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS1
|
ENSG00000143995.15 | Meis homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEIS1 | hg19_v2_chr2_+_66662249_66662267 | 0.99 | 1.2e-02 | Click! |
Activity profile of MEIS1 motif
Sorted Z-values of MEIS1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_150866779 | 1.69 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr17_+_68165657 | 1.44 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr6_+_53794780 | 1.15 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr12_-_57030096 | 0.92 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr2_-_118943930 | 0.64 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr5_+_49962772 | 0.62 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_-_144866711 | 0.60 |
ENST00000530130.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_+_19358192 | 0.58 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr17_-_39341594 | 0.57 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr5_-_138780159 | 0.57 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr16_+_2588012 | 0.53 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr6_-_134861089 | 0.52 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr5_+_49962495 | 0.51 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_-_41111323 | 0.51 |
ENST00000595486.1
|
AL133318.1
|
Uncharacterized protein |
| chr3_-_88108212 | 0.48 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chrX_-_80377162 | 0.48 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr12_+_62654155 | 0.47 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr10_-_977564 | 0.46 |
ENST00000406525.2
|
LARP4B
|
La ribonucleoprotein domain family, member 4B |
| chr5_+_173763250 | 0.45 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr4_+_128886584 | 0.44 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr12_-_42631529 | 0.42 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr17_-_66287310 | 0.42 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr4_-_14889791 | 0.41 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr20_-_35580240 | 0.41 |
ENST00000262878.4
|
SAMHD1
|
SAM domain and HD domain 1 |
| chr17_-_66287257 | 0.40 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr12_+_95611536 | 0.40 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr5_+_122110691 | 0.40 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr16_-_11350036 | 0.39 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr12_+_78224667 | 0.39 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr4_-_101111615 | 0.38 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr5_-_133706695 | 0.38 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr1_-_115301235 | 0.38 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr5_+_149865377 | 0.37 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr8_+_32579271 | 0.37 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr12_+_95612006 | 0.36 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chrX_-_80377118 | 0.36 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr2_+_159651821 | 0.36 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr14_+_52164820 | 0.36 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr16_+_31044812 | 0.36 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr3_-_52719912 | 0.36 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr11_+_64052454 | 0.34 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr11_+_117947724 | 0.34 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr9_-_140142222 | 0.34 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr15_-_33180439 | 0.34 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr8_+_32579321 | 0.33 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr3_-_143567262 | 0.33 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chrX_-_47004437 | 0.33 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr20_-_31172598 | 0.33 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr2_+_37311588 | 0.33 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chrX_+_133930798 | 0.32 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr2_-_239140011 | 0.32 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr16_+_30907927 | 0.32 |
ENST00000279804.2
ENST00000395019.3 |
CTF1
|
cardiotrophin 1 |
| chr4_-_88450372 | 0.32 |
ENST00000543631.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr8_+_86999516 | 0.32 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr2_+_5832799 | 0.31 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr6_+_31105426 | 0.31 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr4_-_130014729 | 0.30 |
ENST00000281142.5
ENST00000434680.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr1_+_27719148 | 0.29 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr17_-_7155775 | 0.28 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr4_+_76871883 | 0.28 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr16_-_3350614 | 0.28 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr12_-_106477805 | 0.28 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr17_-_62502399 | 0.28 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chrX_+_107068959 | 0.28 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr8_-_139926236 | 0.28 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr14_+_91526668 | 0.27 |
ENST00000521334.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chrX_+_107069063 | 0.27 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr20_-_32580924 | 0.27 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr14_+_64970427 | 0.27 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr14_-_55738788 | 0.27 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr17_-_62502639 | 0.27 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr3_-_52719888 | 0.26 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr17_-_46703826 | 0.26 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr10_+_114710516 | 0.26 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_148939598 | 0.26 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_-_94673956 | 0.26 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr11_+_72281681 | 0.26 |
ENST00000450804.3
|
RP11-169D4.1
|
RP11-169D4.1 |
| chr2_-_68547061 | 0.26 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr2_+_24714729 | 0.26 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr10_+_114709999 | 0.26 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_153330322 | 0.26 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr9_-_113341823 | 0.26 |
ENST00000302728.8
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr12_+_57916584 | 0.26 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr10_+_102758105 | 0.26 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr11_+_57308979 | 0.25 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr3_-_182880541 | 0.25 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr17_-_34417479 | 0.25 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr2_+_131769256 | 0.25 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_35198243 | 0.25 |
ENST00000528455.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_222885770 | 0.25 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr13_-_44735393 | 0.25 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr21_+_40823753 | 0.25 |
ENST00000333634.4
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr9_-_16727978 | 0.25 |
ENST00000418777.1
ENST00000468187.2 |
BNC2
|
basonuclin 2 |
| chr9_-_79307096 | 0.25 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr5_+_32712363 | 0.25 |
ENST00000507141.1
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr2_+_218933972 | 0.24 |
ENST00000374155.3
|
RUFY4
|
RUN and FYVE domain containing 4 |
| chr10_+_102672712 | 0.24 |
ENST00000370271.3
ENST00000370269.3 ENST00000609386.1 |
FAM178A
|
family with sequence similarity 178, member A |
| chr18_+_3451584 | 0.24 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_37311445 | 0.24 |
ENST00000233099.5
ENST00000354531.2 |
HEATR5B
|
HEAT repeat containing 5B |
| chr8_-_8318847 | 0.24 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr17_+_21188012 | 0.24 |
ENST00000529517.1
|
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr12_+_113344582 | 0.24 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_+_16080659 | 0.24 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr1_+_161692449 | 0.24 |
ENST00000367946.3
ENST00000367945.1 ENST00000336830.5 ENST00000367944.3 ENST00000392158.1 |
FCRLB
|
Fc receptor-like B |
| chr1_-_117021430 | 0.24 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr2_-_158182105 | 0.24 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr11_-_22851367 | 0.24 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chr13_-_52027134 | 0.24 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr17_-_66287350 | 0.23 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr21_+_35553045 | 0.23 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr9_-_4299874 | 0.23 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr9_+_111696664 | 0.23 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr5_+_135170331 | 0.23 |
ENST00000425402.1
ENST00000274513.5 ENST00000420621.1 ENST00000433282.2 ENST00000412661.2 |
SLC25A48
|
solute carrier family 25, member 48 |
| chr10_-_61513146 | 0.23 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chrX_-_83757399 | 0.23 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr2_+_153191706 | 0.23 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr1_+_100111580 | 0.23 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr3_+_184016986 | 0.22 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr6_+_19837592 | 0.22 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr6_+_142623758 | 0.22 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr3_-_169587621 | 0.21 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr16_+_2587998 | 0.21 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr9_-_100459639 | 0.21 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr2_-_158182322 | 0.21 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr16_+_56691911 | 0.21 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr19_-_46145696 | 0.21 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr8_+_23386557 | 0.21 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr12_+_75874580 | 0.21 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr4_+_141445333 | 0.21 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr14_-_21491305 | 0.20 |
ENST00000554531.1
|
NDRG2
|
NDRG family member 2 |
| chr14_+_45464658 | 0.20 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chr11_-_108338218 | 0.20 |
ENST00000525729.1
ENST00000393084.1 |
C11orf65
|
chromosome 11 open reading frame 65 |
| chr12_-_110937351 | 0.20 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr7_+_128864848 | 0.20 |
ENST00000325006.3
ENST00000446544.2 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr13_-_50510622 | 0.20 |
ENST00000378195.2
|
SPRYD7
|
SPRY domain containing 7 |
| chrX_+_102024075 | 0.20 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr10_-_14050522 | 0.20 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr16_+_30194916 | 0.20 |
ENST00000570045.1
ENST00000565497.1 ENST00000570244.1 |
CORO1A
|
coronin, actin binding protein, 1A |
| chr8_+_38239882 | 0.20 |
ENST00000607047.1
|
RP11-350N15.5
|
RP11-350N15.5 |
| chr1_+_6845384 | 0.20 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr11_+_61717535 | 0.20 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr13_-_50510434 | 0.20 |
ENST00000361840.3
|
SPRYD7
|
SPRY domain containing 7 |
| chrX_+_134654540 | 0.19 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chrX_+_135251783 | 0.19 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr7_-_100171270 | 0.19 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr1_+_100598691 | 0.19 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr3_-_187455680 | 0.19 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr12_+_111843749 | 0.19 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr7_-_32111009 | 0.19 |
ENST00000396184.3
ENST00000396189.2 ENST00000321453.7 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr2_+_54684327 | 0.19 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr1_+_180875711 | 0.19 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr19_-_38743878 | 0.19 |
ENST00000587515.1
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr3_-_161089289 | 0.19 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chrX_+_17393543 | 0.19 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr4_-_159080806 | 0.19 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr4_-_165305086 | 0.19 |
ENST00000507270.1
ENST00000514618.1 ENST00000503008.1 |
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_+_156863470 | 0.19 |
ENST00000338302.3
ENST00000455314.1 ENST00000292357.7 |
PEAR1
|
platelet endothelial aggregation receptor 1 |
| chr22_+_36784632 | 0.18 |
ENST00000424761.1
|
RP4-633O19__A.1
|
RP4-633O19__A.1 |
| chr19_+_21203426 | 0.18 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr4_+_77870960 | 0.18 |
ENST00000505788.1
ENST00000510515.1 ENST00000504637.1 |
SEPT11
|
septin 11 |
| chr18_-_21891460 | 0.18 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr4_+_77941685 | 0.18 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr7_-_127032363 | 0.18 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr5_-_159766528 | 0.18 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr17_-_62502022 | 0.18 |
ENST00000578804.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr16_+_57702099 | 0.18 |
ENST00000333493.4
ENST00000327655.6 |
GPR97
|
G protein-coupled receptor 97 |
| chr10_-_11574274 | 0.18 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr6_-_154751629 | 0.18 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr12_+_128399965 | 0.18 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr13_+_76378305 | 0.18 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr7_-_27169801 | 0.18 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr19_-_52511334 | 0.18 |
ENST00000602063.1
ENST00000597747.1 ENST00000594083.1 ENST00000593650.1 ENST00000599631.1 ENST00000598071.1 ENST00000601178.1 ENST00000376716.5 ENST00000391795.3 |
ZNF615
|
zinc finger protein 615 |
| chr17_+_41857793 | 0.18 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr1_+_104068562 | 0.18 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr17_+_18855470 | 0.18 |
ENST00000395647.2
ENST00000395642.1 ENST00000417251.2 ENST00000395643.2 ENST00000395645.3 |
SLC5A10
|
solute carrier family 5 (sodium/sugar cotransporter), member 10 |
| chr18_-_54318353 | 0.18 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr15_-_71407833 | 0.18 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr12_+_8234807 | 0.17 |
ENST00000339754.5
|
NECAP1
|
NECAP endocytosis associated 1 |
| chr1_+_26605618 | 0.17 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr5_-_175843524 | 0.17 |
ENST00000502877.1
|
CLTB
|
clathrin, light chain B |
| chr9_+_6758024 | 0.17 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr1_-_35497283 | 0.17 |
ENST00000373333.1
|
ZMYM6
|
zinc finger, MYM-type 6 |
| chr12_+_75874460 | 0.17 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr15_+_58724184 | 0.17 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr1_+_228395755 | 0.17 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr7_-_141957847 | 0.17 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chrX_+_49644470 | 0.17 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr13_+_50018402 | 0.17 |
ENST00000354234.4
|
SETDB2
|
SET domain, bifurcated 2 |
| chr1_+_6845497 | 0.17 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr6_+_143772060 | 0.17 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr14_-_71107921 | 0.17 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr7_+_95401877 | 0.17 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr14_-_91526922 | 0.17 |
ENST00000418736.2
ENST00000261991.3 |
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr1_-_243418650 | 0.17 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr11_-_26743546 | 0.17 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr7_+_26438187 | 0.16 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr14_-_58893832 | 0.16 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_-_120365866 | 0.16 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr5_+_133706865 | 0.16 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr10_-_113943447 | 0.16 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.3 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.5 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.3 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.8 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.3 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.6 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.7 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.3 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.2 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 1.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.3 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.1 | 0.2 | GO:0003168 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.3 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:0012502 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 1.0 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:1902728 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.0 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.9 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.2 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.2 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.2 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.1 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.2 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.1 | GO:0045360 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.2 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0051495 | positive regulation of cytoskeleton organization(GO:0051495) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0044727 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.0 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:1903276 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 1.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 1.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.6 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.9 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.3 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.2 | GO:0050659 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0015265 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |