Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
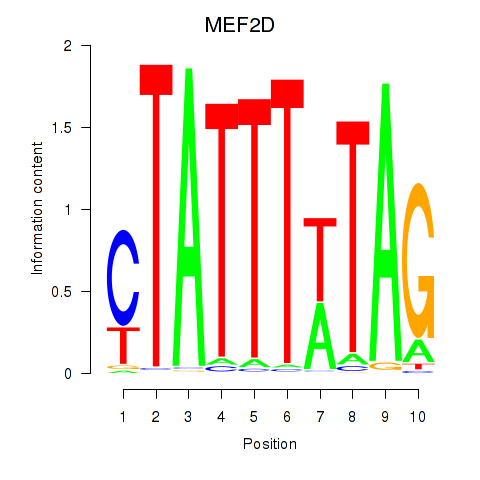
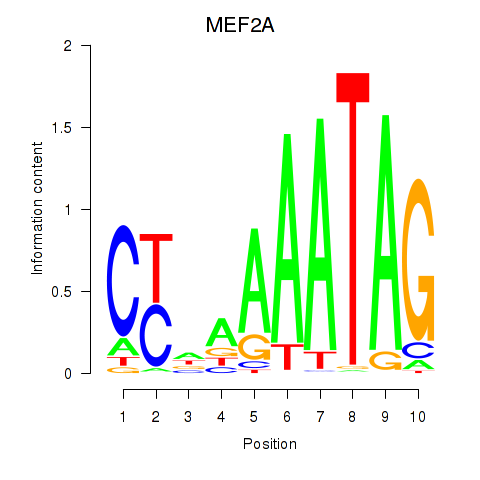
Results for MEF2D_MEF2A
Z-value: 0.56
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2A | hg19_v2_chr15_+_100106155_100106243 | 0.73 | 2.7e-01 | Click! |
| MEF2D | hg19_v2_chr1_-_156470556_156470643 | -0.66 | 3.4e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34417479 | 1.78 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr11_+_57308979 | 1.20 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr7_-_134143841 | 0.93 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr11_+_12108410 | 0.92 |
ENST00000527997.1
|
RP13-631K18.5
|
RP13-631K18.5 |
| chr3_-_108248169 | 0.64 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr8_+_107282389 | 0.57 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr1_-_100643765 | 0.49 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr8_+_107282368 | 0.47 |
ENST00000521369.2
|
RP11-395G23.3
|
RP11-395G23.3 |
| chr17_+_68165657 | 0.46 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr17_+_7758374 | 0.44 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr17_+_80186908 | 0.43 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_48598547 | 0.41 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr2_+_220299547 | 0.39 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr2_+_233497931 | 0.37 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr6_+_26020672 | 0.37 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr3_+_138067521 | 0.37 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr3_-_156878482 | 0.36 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr2_-_145188137 | 0.35 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_23282797 | 0.34 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr6_-_154751629 | 0.33 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr14_-_62217779 | 0.33 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr22_-_51017084 | 0.31 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr2_+_33661382 | 0.30 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr8_-_23282820 | 0.30 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr22_-_51016846 | 0.29 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr17_-_6917755 | 0.28 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr7_-_41742697 | 0.27 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr15_+_100106155 | 0.26 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr11_+_65082289 | 0.26 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr22_-_30642728 | 0.25 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr11_-_104480019 | 0.25 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr1_+_209878182 | 0.25 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr8_+_40010989 | 0.25 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr9_+_5450503 | 0.24 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr20_+_48807351 | 0.23 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr2_+_33683109 | 0.23 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_218766698 | 0.23 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr15_+_100106126 | 0.23 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr14_+_103589789 | 0.23 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr3_+_138067666 | 0.23 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr3_-_156878540 | 0.22 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr6_-_26124138 | 0.22 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr5_-_141704566 | 0.22 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr19_-_47128294 | 0.21 |
ENST00000596260.1
ENST00000597185.1 ENST00000598865.1 ENST00000594275.1 ENST00000291294.2 |
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr1_-_12677714 | 0.21 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr8_-_70745575 | 0.21 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr2_-_165424973 | 0.21 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr17_+_7184986 | 0.20 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr2_-_31637560 | 0.20 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chrX_+_18725758 | 0.19 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr9_+_100263912 | 0.19 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr9_-_136344237 | 0.19 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr9_+_127054254 | 0.17 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr2_-_208030647 | 0.17 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr17_-_34524157 | 0.17 |
ENST00000378354.4
ENST00000394484.1 |
CCL3L3
|
chemokine (C-C motif) ligand 3-like 3 |
| chr8_-_62602327 | 0.17 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr7_+_18536090 | 0.17 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr7_+_28448995 | 0.17 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr3_+_138067314 | 0.17 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr6_+_15401075 | 0.16 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr7_-_20826504 | 0.16 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr10_+_102758105 | 0.16 |
ENST00000429732.1
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chrX_+_591524 | 0.16 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr17_-_48277552 | 0.15 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr12_-_16760195 | 0.15 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr6_+_131571535 | 0.15 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr9_-_136344197 | 0.15 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr6_-_6007200 | 0.15 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr4_-_185303418 | 0.15 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr18_-_3220106 | 0.15 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr3_-_137834436 | 0.15 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr1_-_59249732 | 0.14 |
ENST00000371222.2
|
JUN
|
jun proto-oncogene |
| chr18_+_3252206 | 0.14 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_-_917497 | 0.14 |
ENST00000433179.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr10_+_71561630 | 0.14 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_-_106054952 | 0.14 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr15_+_74509530 | 0.14 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr11_+_3666335 | 0.14 |
ENST00000250693.1
|
ART1
|
ADP-ribosyltransferase 1 |
| chr16_+_31225337 | 0.14 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr22_+_31489344 | 0.14 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr1_-_201391149 | 0.14 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr3_-_4927447 | 0.13 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr2_+_113403434 | 0.13 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr20_+_42574317 | 0.13 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr8_+_1993152 | 0.13 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr1_+_228395755 | 0.13 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr8_-_73793975 | 0.13 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr4_+_57276661 | 0.13 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr19_+_35629702 | 0.13 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr15_+_100106244 | 0.13 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr9_+_127023704 | 0.12 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr19_-_47975417 | 0.12 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr4_+_41362796 | 0.12 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_-_27949911 | 0.12 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr1_+_25757376 | 0.12 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr1_-_223853348 | 0.12 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr5_+_155753745 | 0.12 |
ENST00000435422.3
ENST00000337851.4 ENST00000447401.1 |
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr1_-_25756638 | 0.12 |
ENST00000349320.3
|
RHCE
|
Rh blood group, CcEe antigens |
| chr4_+_160203650 | 0.12 |
ENST00000514565.1
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_47293795 | 0.12 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr2_-_208031542 | 0.12 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_201346761 | 0.12 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr18_-_35145981 | 0.12 |
ENST00000420428.2
ENST00000412753.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr6_-_169654139 | 0.11 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr18_+_9708162 | 0.11 |
ENST00000578921.1
|
RAB31
|
RAB31, member RAS oncogene family |
| chr15_+_91446961 | 0.11 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr9_+_21440440 | 0.11 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr18_-_3219847 | 0.11 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr14_-_103989033 | 0.11 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chrX_-_55208866 | 0.11 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr1_-_201390846 | 0.11 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr1_-_116311402 | 0.11 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chrX_-_133792480 | 0.11 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chrM_+_7586 | 0.11 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_+_75931861 | 0.10 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr22_-_31328881 | 0.10 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr3_-_145878954 | 0.10 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr2_+_66918558 | 0.10 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr4_-_23891658 | 0.10 |
ENST00000507380.1
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr8_+_95565947 | 0.10 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr8_+_1993173 | 0.10 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr6_+_138266498 | 0.10 |
ENST00000434437.1
ENST00000417800.1 |
RP11-240M16.1
|
RP11-240M16.1 |
| chr13_+_76378407 | 0.10 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr1_-_917466 | 0.10 |
ENST00000341290.2
|
C1orf170
|
chromosome 1 open reading frame 170 |
| chr8_-_42234745 | 0.10 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr7_-_124569864 | 0.10 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr10_+_77056181 | 0.10 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chr3_-_157823839 | 0.10 |
ENST00000425436.3
ENST00000389589.4 ENST00000441443.2 |
SHOX2
|
short stature homeobox 2 |
| chr7_+_120629653 | 0.10 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr20_+_9049742 | 0.10 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr12_+_95612006 | 0.10 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr7_+_47694842 | 0.10 |
ENST00000408988.2
|
C7orf65
|
chromosome 7 open reading frame 65 |
| chr12_-_125401885 | 0.09 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr2_+_179149636 | 0.09 |
ENST00000409631.1
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr6_+_35996859 | 0.09 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr17_+_27369918 | 0.09 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr3_+_35683651 | 0.09 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr3_+_121311966 | 0.09 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr12_+_109554386 | 0.09 |
ENST00000338432.7
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr20_+_36373032 | 0.09 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr7_-_143582450 | 0.09 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr3_-_157824292 | 0.09 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr2_+_65663812 | 0.09 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr15_+_80364901 | 0.09 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr18_+_3252265 | 0.09 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr13_+_76334795 | 0.09 |
ENST00000526202.1
ENST00000465261.2 |
LMO7
|
LIM domain 7 |
| chr20_+_61147651 | 0.09 |
ENST00000370527.3
ENST00000370524.2 |
C20orf166
|
chromosome 20 open reading frame 166 |
| chr17_-_39211463 | 0.09 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr13_-_46756351 | 0.09 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr10_-_75415825 | 0.09 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr16_-_3422283 | 0.09 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr19_+_33071974 | 0.09 |
ENST00000590247.2
ENST00000419343.3 ENST00000592786.1 ENST00000379316.3 |
PDCD5
|
programmed cell death 5 |
| chr12_+_119616447 | 0.08 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr12_+_12938541 | 0.08 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr4_-_44653636 | 0.08 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr9_+_8858102 | 0.08 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chrX_-_15332665 | 0.08 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr22_-_37215523 | 0.08 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr10_-_49860525 | 0.08 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr19_+_49927006 | 0.08 |
ENST00000576655.1
|
CTD-3148I10.1
|
golgi-associated, olfactory signaling regulator |
| chr10_-_95241951 | 0.08 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr8_-_49833978 | 0.08 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr9_+_127054217 | 0.08 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr22_+_41487711 | 0.08 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr11_+_1860200 | 0.08 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr1_+_84630574 | 0.07 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr20_+_1875110 | 0.07 |
ENST00000400068.3
|
SIRPA
|
signal-regulatory protein alpha |
| chr5_-_115910630 | 0.07 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_95242044 | 0.07 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr4_+_55095264 | 0.07 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr10_-_75410771 | 0.07 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr8_+_27182862 | 0.07 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_+_182584314 | 0.07 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr3_+_127634312 | 0.07 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr2_+_220144052 | 0.07 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr15_+_65369082 | 0.07 |
ENST00000432196.2
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr5_-_137475071 | 0.07 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr18_-_64271316 | 0.07 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr6_+_132129151 | 0.07 |
ENST00000360971.2
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr7_-_151217001 | 0.07 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr10_+_71562180 | 0.07 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr18_-_35145728 | 0.07 |
ENST00000361795.5
ENST00000603232.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr3_+_135741576 | 0.07 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_+_74701062 | 0.07 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_+_170633047 | 0.07 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr21_-_28215332 | 0.07 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr11_+_128563652 | 0.07 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_+_115358088 | 0.07 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr13_+_76334567 | 0.07 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr10_+_71561649 | 0.07 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_-_122247879 | 0.07 |
ENST00000545861.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr6_+_53948328 | 0.07 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr12_+_14561422 | 0.07 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_179184955 | 0.07 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr1_+_1846519 | 0.07 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr10_+_50507232 | 0.07 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr2_+_30569506 | 0.07 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr12_-_102874102 | 0.06 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chrX_+_9217932 | 0.06 |
ENST00000432442.1
|
GS1-519E5.1
|
GS1-519E5.1 |
| chr19_-_47975143 | 0.06 |
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.3 | 1.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 0.6 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 1.2 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.6 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:0071288 | carbon dioxide transmembrane transport(GO:0035378) positive regulation of saliva secretion(GO:0046878) cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.0 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.0 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |