Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
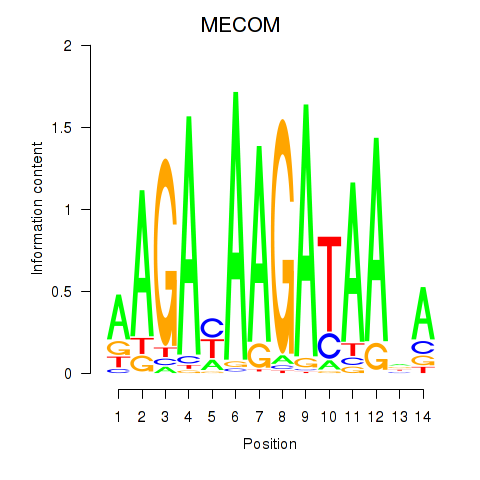
Results for MECOM
Z-value: 0.84
Transcription factors associated with MECOM
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MECOM
|
ENSG00000085276.13 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MECOM | hg19_v2_chr3_-_168864427_168864464 | 0.72 | 2.8e-01 | Click! |
Activity profile of MECOM motif
Sorted Z-values of MECOM motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10320190 | 0.60 |
ENST00000543993.1
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr8_+_58658679 | 0.52 |
ENST00000518934.1
|
CTD-2339F6.1
|
CTD-2339F6.1 |
| chr8_+_86999516 | 0.52 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr3_-_96337000 | 0.48 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr11_+_17316870 | 0.45 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr1_+_161035655 | 0.36 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr15_-_102285007 | 0.36 |
ENST00000560292.2
|
RP11-89K11.1
|
Uncharacterized protein |
| chr5_-_79946820 | 0.35 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr1_-_238108575 | 0.34 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr13_-_46626820 | 0.34 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr12_+_124457746 | 0.34 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr2_+_242289502 | 0.32 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr12_-_8218997 | 0.31 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr8_-_133637624 | 0.31 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr21_+_17214724 | 0.30 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr1_-_179834311 | 0.30 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr12_-_112443830 | 0.29 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr9_+_26746951 | 0.29 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr6_+_49431073 | 0.29 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr15_-_37391507 | 0.28 |
ENST00000557796.2
ENST00000397620.2 |
MEIS2
|
Meis homeobox 2 |
| chr6_+_28911654 | 0.27 |
ENST00000377186.3
|
C6orf100
|
chromosome 6 open reading frame 100 |
| chr14_-_39639523 | 0.26 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr18_-_21891460 | 0.26 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr15_+_100348193 | 0.26 |
ENST00000558188.1
|
CTD-2054N24.2
|
Uncharacterized protein |
| chr3_-_119379427 | 0.25 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr3_-_45883558 | 0.25 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chrX_-_149999722 | 0.25 |
ENST00000418547.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr11_+_125703204 | 0.24 |
ENST00000534411.1
ENST00000457514.2 |
PATE4
|
prostate and testis expressed 4 |
| chr3_-_145968923 | 0.24 |
ENST00000493382.1
ENST00000354952.2 ENST00000383083.2 |
PLSCR4
|
phospholipid scramblase 4 |
| chr4_-_76944621 | 0.23 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr12_+_28605426 | 0.23 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr10_+_116524547 | 0.21 |
ENST00000436932.1
|
RP11-106M7.1
|
RP11-106M7.1 |
| chr3_-_130746462 | 0.21 |
ENST00000505545.1
|
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr3_-_119379719 | 0.21 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr5_+_159895275 | 0.20 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr6_+_27215494 | 0.20 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr8_-_90993869 | 0.20 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr5_+_102200948 | 0.20 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_+_99986036 | 0.20 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr22_-_30866564 | 0.19 |
ENST00000435069.1
ENST00000415957.2 ENST00000540910.1 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr12_+_96588368 | 0.19 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr7_+_142374104 | 0.19 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr15_+_40532723 | 0.19 |
ENST00000558878.1
ENST00000558183.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr4_+_155484155 | 0.18 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr19_-_9785743 | 0.18 |
ENST00000537617.1
ENST00000589542.1 ENST00000590155.1 ENST00000541032.1 ENST00000588653.1 ENST00000448622.1 ENST00000453792.2 |
ZNF562
|
zinc finger protein 562 |
| chr2_+_27805880 | 0.18 |
ENST00000379717.1
ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512
|
zinc finger protein 512 |
| chr3_-_112738565 | 0.17 |
ENST00000383675.2
ENST00000314400.5 |
C3orf17
|
chromosome 3 open reading frame 17 |
| chr6_-_52860171 | 0.17 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr6_+_27215471 | 0.17 |
ENST00000421826.2
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr19_-_54106751 | 0.17 |
ENST00000600193.1
|
CTB-167G5.5
|
Uncharacterized protein |
| chr5_+_64920543 | 0.16 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr4_+_155484103 | 0.16 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr7_-_100844193 | 0.16 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr12_-_86650077 | 0.16 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_-_83837983 | 0.16 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr1_+_40810516 | 0.16 |
ENST00000435168.2
|
SMAP2
|
small ArfGAP2 |
| chr8_-_95274536 | 0.16 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr4_-_52883786 | 0.15 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr14_+_73563735 | 0.15 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr8_-_80993010 | 0.15 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr5_-_64920115 | 0.15 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr5_+_56471592 | 0.15 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr9_-_13175823 | 0.14 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_-_145275109 | 0.14 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_56492816 | 0.14 |
ENST00000522360.1
|
DST
|
dystonin |
| chr9_+_33240157 | 0.14 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chrX_+_155227246 | 0.14 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr4_-_88450372 | 0.14 |
ENST00000543631.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr2_-_145275828 | 0.14 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_-_85302531 | 0.14 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr2_-_70409953 | 0.13 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chrX_+_66764375 | 0.13 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr13_+_27844464 | 0.13 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr7_+_17338239 | 0.13 |
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor |
| chr13_+_76334498 | 0.13 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr10_+_106034637 | 0.13 |
ENST00000401888.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr12_+_21168630 | 0.13 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr6_-_53530474 | 0.13 |
ENST00000370905.3
|
KLHL31
|
kelch-like family member 31 |
| chr2_-_56150910 | 0.13 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr17_+_53344945 | 0.12 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr16_+_9449445 | 0.12 |
ENST00000564305.1
|
RP11-243A14.1
|
RP11-243A14.1 |
| chr2_-_224467093 | 0.12 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr12_+_72080253 | 0.12 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr21_+_45595354 | 0.12 |
ENST00000411956.1
|
AP001056.1
|
AP001056.1 |
| chr3_-_184971853 | 0.12 |
ENST00000231887.3
|
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr8_-_27115903 | 0.11 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr2_-_171571077 | 0.11 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr3_-_161089289 | 0.11 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr3_-_145968857 | 0.11 |
ENST00000433593.2
ENST00000476202.1 ENST00000460885.1 |
PLSCR4
|
phospholipid scramblase 4 |
| chr3_+_101659682 | 0.11 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr2_+_204571198 | 0.11 |
ENST00000374481.3
ENST00000458610.2 ENST00000324106.8 |
CD28
|
CD28 molecule |
| chr5_+_67485704 | 0.11 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr12_+_32638897 | 0.11 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_218683438 | 0.11 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr11_-_33891362 | 0.11 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr5_+_92919043 | 0.11 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr17_+_26638667 | 0.11 |
ENST00000600595.1
|
AC061975.10
|
Uncharacterized protein |
| chr15_-_48470544 | 0.10 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr15_-_64673630 | 0.10 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr4_+_117220016 | 0.10 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr11_-_73689037 | 0.10 |
ENST00000544615.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr13_+_108870714 | 0.10 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr4_-_99851766 | 0.10 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr3_-_109035342 | 0.10 |
ENST00000478945.1
|
DPPA2
|
developmental pluripotency associated 2 |
| chr2_-_157189180 | 0.10 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_+_25664408 | 0.10 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr15_+_52155001 | 0.10 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr12_+_20848377 | 0.09 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_96588279 | 0.09 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr10_-_18940501 | 0.09 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr1_+_110158726 | 0.09 |
ENST00000531734.1
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr10_-_14596140 | 0.09 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr1_+_160160346 | 0.09 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr16_-_73093597 | 0.09 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr7_-_33140498 | 0.09 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr2_+_190541153 | 0.09 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr13_+_111855414 | 0.09 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr14_+_24584372 | 0.09 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr17_-_32484313 | 0.09 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr6_-_132022635 | 0.08 |
ENST00000315453.2
|
OR2A4
|
olfactory receptor, family 2, subfamily A, member 4 |
| chr1_+_82266053 | 0.08 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr1_+_160160283 | 0.08 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr14_+_24584056 | 0.08 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_12115543 | 0.08 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr15_-_55562451 | 0.08 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr16_-_50402836 | 0.08 |
ENST00000394688.3
|
BRD7
|
bromodomain containing 7 |
| chr2_+_55746746 | 0.08 |
ENST00000406691.3
ENST00000349456.4 ENST00000407816.3 ENST00000403007.3 |
CCDC104
|
coiled-coil domain containing 104 |
| chr1_-_202927490 | 0.07 |
ENST00000340990.5
|
ADIPOR1
|
adiponectin receptor 1 |
| chr16_+_31225337 | 0.07 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr8_+_11351494 | 0.07 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr12_-_15865844 | 0.07 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr2_+_97779233 | 0.07 |
ENST00000461153.2
ENST00000420699.2 |
ANKRD36
|
ankyrin repeat domain 36 |
| chr2_+_55746722 | 0.07 |
ENST00000339012.3
|
CCDC104
|
coiled-coil domain containing 104 |
| chr5_+_96211643 | 0.07 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr19_+_22235279 | 0.07 |
ENST00000594363.1
ENST00000597927.1 ENST00000594947.1 |
ZNF257
|
zinc finger protein 257 |
| chr2_+_37571717 | 0.07 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr2_-_1629176 | 0.07 |
ENST00000366424.2
|
AC144450.2
|
AC144450.2 |
| chr4_-_77997126 | 0.07 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr14_-_69261310 | 0.07 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr12_-_371994 | 0.07 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr10_-_75168071 | 0.07 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr4_-_122148620 | 0.07 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr15_+_71185148 | 0.07 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr19_-_22966893 | 0.07 |
ENST00000596209.1
|
ZNF99
|
zinc finger protein 99 |
| chr15_+_71839566 | 0.07 |
ENST00000357769.4
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr1_+_248031277 | 0.07 |
ENST00000537741.1
|
OR2W3
|
olfactory receptor, family 2, subfamily W, member 3 |
| chr12_+_72079842 | 0.06 |
ENST00000266673.5
ENST00000550524.1 |
TMEM19
|
transmembrane protein 19 |
| chr6_-_130543958 | 0.06 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr2_-_145275211 | 0.06 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_22022437 | 0.06 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr12_-_121973974 | 0.06 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr12_-_85430024 | 0.06 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr1_-_40041925 | 0.06 |
ENST00000372862.3
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr8_+_105602961 | 0.06 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr8_+_11351876 | 0.06 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr10_+_57358750 | 0.06 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr1_-_68962805 | 0.06 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chrY_+_15418467 | 0.06 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr5_+_140165876 | 0.06 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr11_-_10530723 | 0.06 |
ENST00000536684.1
|
MTRNR2L8
|
MT-RNR2-like 8 |
| chr2_+_190540711 | 0.06 |
ENST00000520309.1
|
ANKAR
|
ankyrin and armadillo repeat containing |
| chr3_-_145968900 | 0.05 |
ENST00000460350.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr7_-_76039000 | 0.05 |
ENST00000275560.3
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains) |
| chr5_-_180235755 | 0.05 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr4_+_129349188 | 0.05 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr14_-_75179774 | 0.05 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr2_+_27805971 | 0.05 |
ENST00000413371.2
|
ZNF512
|
zinc finger protein 512 |
| chr12_-_39734783 | 0.05 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr17_-_43138357 | 0.05 |
ENST00000342350.5
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr5_-_171404730 | 0.05 |
ENST00000518752.1
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr12_-_11339543 | 0.05 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr3_-_149093499 | 0.05 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr10_+_38717074 | 0.05 |
ENST00000423687.1
|
LINC00999
|
long intergenic non-protein coding RNA 999 |
| chr5_-_133340682 | 0.05 |
ENST00000265333.3
|
VDAC1
|
voltage-dependent anion channel 1 |
| chr14_-_24551137 | 0.05 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chrX_+_86772787 | 0.05 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chrX_-_106243451 | 0.05 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chrX_-_43741594 | 0.04 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr14_-_24584138 | 0.04 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr7_-_8302207 | 0.04 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr14_+_19377522 | 0.04 |
ENST00000550708.1
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12 |
| chr3_-_178984759 | 0.04 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr14_-_76447336 | 0.04 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor, beta 3 |
| chr15_-_30686052 | 0.04 |
ENST00000562729.1
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr7_-_15601595 | 0.04 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr6_-_52859968 | 0.04 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr14_-_102701740 | 0.04 |
ENST00000561150.1
ENST00000522867.1 |
MOK
|
MOK protein kinase |
| chr6_+_89674246 | 0.04 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr3_-_184971817 | 0.04 |
ENST00000440662.1
ENST00000456310.1 |
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr16_+_28943260 | 0.03 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr2_+_172544182 | 0.03 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_-_9913489 | 0.03 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr12_+_80838126 | 0.03 |
ENST00000266688.5
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q |
| chr4_+_164415785 | 0.03 |
ENST00000513272.1
ENST00000513134.1 |
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr10_-_48806939 | 0.03 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr19_-_51611623 | 0.03 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr20_+_15177480 | 0.03 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr13_+_76334567 | 0.03 |
ENST00000321797.8
|
LMO7
|
LIM domain 7 |
| chr13_+_31309645 | 0.03 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr14_+_64680854 | 0.03 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MECOM
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |