Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
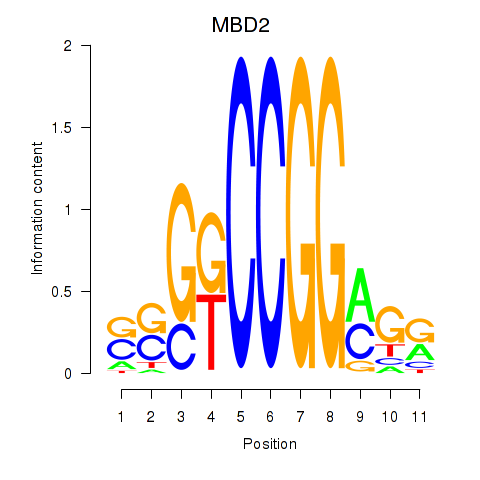
Results for MBD2
Z-value: 1.73
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MBD2 | hg19_v2_chr18_-_51750948_51751059 | 0.78 | 2.2e-01 | Click! |
Activity profile of MBD2 motif
Sorted Z-values of MBD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_51750948 | 1.13 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr6_-_43596899 | 1.02 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr6_+_29795595 | 0.95 |
ENST00000360323.6
ENST00000376818.3 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr17_+_68165657 | 0.93 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr5_+_14143728 | 0.87 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr2_+_220299547 | 0.79 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr2_+_5832799 | 0.76 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr16_+_2059872 | 0.76 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr22_+_19419425 | 0.73 |
ENST00000333130.3
|
MRPL40
|
mitochondrial ribosomal protein L40 |
| chr1_+_3689325 | 0.70 |
ENST00000444870.2
ENST00000452264.1 |
SMIM1
|
small integral membrane protein 1 (Vel blood group) |
| chr19_+_56152262 | 0.66 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr3_-_128206759 | 0.65 |
ENST00000430265.2
|
GATA2
|
GATA binding protein 2 |
| chr20_-_56284816 | 0.65 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr8_-_145688231 | 0.63 |
ENST00000530374.1
|
CYHR1
|
cysteine/histidine-rich 1 |
| chr6_+_17281573 | 0.61 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr4_-_78740511 | 0.61 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr16_-_51185149 | 0.60 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr2_+_220306745 | 0.59 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr18_+_72265084 | 0.57 |
ENST00000582337.1
|
ZNF407
|
zinc finger protein 407 |
| chr1_+_156698743 | 0.57 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr17_+_64961026 | 0.57 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr9_+_137967268 | 0.56 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr19_-_14201507 | 0.56 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr17_+_48638371 | 0.55 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr17_+_65373531 | 0.54 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr8_-_57123815 | 0.54 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr2_+_176972000 | 0.53 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr17_-_42276574 | 0.53 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr19_+_54412517 | 0.52 |
ENST00000391767.1
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7 |
| chr20_-_61885826 | 0.52 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr12_-_107168696 | 0.52 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr17_+_48133459 | 0.51 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr7_-_8301869 | 0.51 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr12_+_122064673 | 0.49 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr11_+_64685026 | 0.49 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr5_-_176778523 | 0.49 |
ENST00000513877.1
ENST00000515209.1 ENST00000514458.1 ENST00000502560.1 |
LMAN2
|
lectin, mannose-binding 2 |
| chr19_-_4124079 | 0.47 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr6_+_45389893 | 0.47 |
ENST00000371432.3
|
RUNX2
|
runt-related transcription factor 2 |
| chr1_-_154531095 | 0.47 |
ENST00000292211.4
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr17_+_65374075 | 0.47 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_-_100493482 | 0.47 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr16_+_2022036 | 0.47 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr19_-_36606181 | 0.45 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr17_-_27278445 | 0.45 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr15_-_65067773 | 0.43 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
| chr1_+_43855545 | 0.42 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr5_+_49962772 | 0.42 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr5_-_176778803 | 0.41 |
ENST00000303127.7
|
LMAN2
|
lectin, mannose-binding 2 |
| chr1_-_31712401 | 0.41 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr12_+_105724613 | 0.41 |
ENST00000549934.2
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr11_+_69455855 | 0.41 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr3_+_133292851 | 0.41 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr11_+_61735266 | 0.40 |
ENST00000601917.1
|
AP003733.1
|
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
| chr16_-_2059797 | 0.40 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr7_-_19157248 | 0.40 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr11_-_46142948 | 0.39 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr19_+_4007644 | 0.39 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr19_+_36606933 | 0.39 |
ENST00000586868.1
|
TBCB
|
tubulin folding cofactor B |
| chr5_-_180076580 | 0.39 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr3_-_88108212 | 0.39 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_+_219824357 | 0.38 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr11_-_61735029 | 0.38 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr16_-_2059748 | 0.38 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr9_+_135037334 | 0.37 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr20_-_48532019 | 0.37 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr19_-_19051103 | 0.37 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr19_-_46476791 | 0.36 |
ENST00000263257.5
|
NOVA2
|
neuro-oncological ventral antigen 2 |
| chr16_-_30905584 | 0.36 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr11_-_6633799 | 0.36 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr7_-_8301768 | 0.35 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr3_+_133292759 | 0.35 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr18_+_8609402 | 0.34 |
ENST00000329286.6
|
RAB12
|
RAB12, member RAS oncogene family |
| chr8_-_141645645 | 0.34 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr22_-_39096981 | 0.34 |
ENST00000427389.1
|
JOSD1
|
Josephin domain containing 1 |
| chr1_-_160232312 | 0.34 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr12_-_93835665 | 0.34 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr17_-_80231300 | 0.34 |
ENST00000398519.5
ENST00000580446.1 |
CSNK1D
|
casein kinase 1, delta |
| chr1_+_6845384 | 0.33 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_-_194207388 | 0.33 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr3_-_124774802 | 0.33 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr20_-_48532046 | 0.32 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr11_-_115375107 | 0.32 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr20_-_1447547 | 0.31 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr3_-_196295437 | 0.31 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr5_+_10564432 | 0.31 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr5_+_170846640 | 0.31 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr11_-_61735103 | 0.31 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr15_+_80351910 | 0.31 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr19_+_35491174 | 0.30 |
ENST00000317991.5
ENST00000504615.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr2_+_54198210 | 0.30 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr10_+_75545391 | 0.30 |
ENST00000604524.1
ENST00000605216.1 ENST00000398706.2 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr14_+_53019822 | 0.30 |
ENST00000321662.6
|
GPR137C
|
G protein-coupled receptor 137C |
| chr12_+_121078355 | 0.30 |
ENST00000316803.3
|
CABP1
|
calcium binding protein 1 |
| chr10_+_75545329 | 0.30 |
ENST00000604729.1
ENST00000603114.1 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr3_+_49591881 | 0.29 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr3_-_183979251 | 0.29 |
ENST00000296238.3
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr19_+_6739662 | 0.29 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr19_+_36606654 | 0.29 |
ENST00000588385.1
ENST00000585746.1 |
TBCB
|
tubulin folding cofactor B |
| chr12_+_6875519 | 0.29 |
ENST00000389462.4
ENST00000540874.1 ENST00000309083.6 |
PTMS
|
parathymosin |
| chr19_-_663171 | 0.28 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr11_-_61734599 | 0.28 |
ENST00000532601.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr20_-_1447467 | 0.28 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chrX_+_68725084 | 0.28 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr19_-_17445613 | 0.28 |
ENST00000159087.4
|
ANO8
|
anoctamin 8 |
| chr1_-_43855444 | 0.28 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr2_-_219537134 | 0.28 |
ENST00000295704.2
|
RNF25
|
ring finger protein 25 |
| chr9_-_130889990 | 0.28 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr12_-_58159361 | 0.28 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr5_-_180076613 | 0.28 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chr9_+_36136700 | 0.27 |
ENST00000396613.3
ENST00000377959.1 ENST00000377960.4 |
GLIPR2
|
GLI pathogenesis-related 2 |
| chr17_-_42441204 | 0.27 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr17_-_80231557 | 0.27 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr8_-_93115445 | 0.27 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_-_31763276 | 0.27 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr15_+_83654950 | 0.27 |
ENST00000304191.3
|
FAM103A1
|
family with sequence similarity 103, member A1 |
| chr14_+_103851712 | 0.27 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr1_+_43148059 | 0.27 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr2_-_220408430 | 0.26 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr11_-_64684672 | 0.26 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr20_+_31350184 | 0.26 |
ENST00000328111.2
ENST00000353855.2 ENST00000348286.2 |
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta |
| chr7_-_8301682 | 0.26 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr16_-_3450963 | 0.26 |
ENST00000573327.1
ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32
|
zinc finger and SCAN domain containing 32 |
| chr19_+_18496957 | 0.26 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr19_+_35491330 | 0.26 |
ENST00000411896.2
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr22_+_19701985 | 0.26 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr3_+_184097905 | 0.26 |
ENST00000450923.1
|
CHRD
|
chordin |
| chr17_-_27278304 | 0.25 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr17_+_43299241 | 0.25 |
ENST00000328118.3
|
FMNL1
|
formin-like 1 |
| chr10_-_11653753 | 0.25 |
ENST00000609104.1
|
USP6NL
|
USP6 N-terminal like |
| chr6_+_1389989 | 0.25 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr20_+_60878005 | 0.25 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr14_-_62162541 | 0.25 |
ENST00000557544.1
|
HIF1A-AS1
|
HIF1A antisense RNA 1 |
| chr19_-_663147 | 0.24 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr15_+_92397051 | 0.24 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr14_-_99947121 | 0.24 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chr1_-_43855479 | 0.24 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr6_+_31865552 | 0.24 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr11_+_67033881 | 0.24 |
ENST00000308595.5
ENST00000526285.1 |
ADRBK1
|
adrenergic, beta, receptor kinase 1 |
| chr17_+_43299156 | 0.23 |
ENST00000331495.3
|
FMNL1
|
formin-like 1 |
| chr20_-_30311703 | 0.23 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr17_-_27949911 | 0.23 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr11_-_62368696 | 0.23 |
ENST00000527204.1
|
MTA2
|
metastasis associated 1 family, member 2 |
| chr11_+_118978045 | 0.23 |
ENST00000336702.3
|
C2CD2L
|
C2CD2-like |
| chr19_+_36606354 | 0.23 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr1_-_32229934 | 0.23 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr11_+_65408273 | 0.23 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr7_+_96634850 | 0.23 |
ENST00000518156.2
|
DLX6
|
distal-less homeobox 6 |
| chr8_-_142318398 | 0.23 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr8_-_119124045 | 0.23 |
ENST00000378204.2
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr8_+_144349606 | 0.23 |
ENST00000521682.1
ENST00000340042.1 |
GLI4
|
GLI family zinc finger 4 |
| chr14_-_99737822 | 0.23 |
ENST00000345514.2
ENST00000443726.2 |
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr1_-_155243235 | 0.23 |
ENST00000355560.4
ENST00000368361.4 |
CLK2
|
CDC-like kinase 2 |
| chr21_+_45875354 | 0.23 |
ENST00000291592.4
|
LRRC3
|
leucine rich repeat containing 3 |
| chr16_-_17564738 | 0.23 |
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I |
| chr3_+_112930373 | 0.23 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr11_-_46142615 | 0.22 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr14_+_102829381 | 0.22 |
ENST00000558678.1
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr2_+_149402553 | 0.22 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr2_+_74212073 | 0.22 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr16_-_30022293 | 0.22 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr14_-_99947168 | 0.22 |
ENST00000331768.5
|
SETD3
|
SET domain containing 3 |
| chr3_+_112930946 | 0.22 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr11_+_64851666 | 0.22 |
ENST00000525509.1
ENST00000294258.3 ENST00000526334.1 |
ZFPL1
|
zinc finger protein-like 1 |
| chr20_+_34894247 | 0.22 |
ENST00000373913.3
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr7_+_69064566 | 0.22 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr16_+_3019246 | 0.22 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr2_-_208031542 | 0.21 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_+_21538517 | 0.21 |
ENST00000298693.3
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr19_-_17137625 | 0.21 |
ENST00000443236.1
ENST00000388925.4 |
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr14_-_103523745 | 0.21 |
ENST00000361246.2
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr22_+_19702069 | 0.21 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr17_-_46690839 | 0.21 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chrX_+_12156582 | 0.21 |
ENST00000380682.1
|
FRMPD4
|
FERM and PDZ domain containing 4 |
| chr1_-_32229523 | 0.21 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr16_-_28074822 | 0.21 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr12_+_110719032 | 0.20 |
ENST00000395494.2
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr14_+_93799556 | 0.20 |
ENST00000256339.4
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr1_+_43855560 | 0.20 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr18_-_53255766 | 0.20 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr7_-_127671674 | 0.20 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr11_+_66025938 | 0.20 |
ENST00000394066.2
|
KLC2
|
kinesin light chain 2 |
| chr12_-_124457371 | 0.20 |
ENST00000238156.3
ENST00000545037.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr17_-_7142725 | 0.20 |
ENST00000571362.1
ENST00000576955.1 ENST00000320316.3 |
PHF23
|
PHD finger protein 23 |
| chr9_-_74383799 | 0.20 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr11_+_67056755 | 0.19 |
ENST00000511455.2
ENST00000308440.6 |
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr3_+_14989076 | 0.19 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr6_-_42981651 | 0.19 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr12_+_57482665 | 0.19 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr1_+_43148625 | 0.19 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr3_+_33155525 | 0.19 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr12_+_132195617 | 0.19 |
ENST00000261674.4
ENST00000535236.1 ENST00000541286.1 |
SFSWAP
|
splicing factor, suppressor of white-apricot homolog (Drosophila) |
| chr16_-_89787360 | 0.19 |
ENST00000389386.3
|
VPS9D1
|
VPS9 domain containing 1 |
| chr4_-_11430221 | 0.19 |
ENST00000514690.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr7_+_128470431 | 0.19 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chr11_+_67055986 | 0.19 |
ENST00000447274.2
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr13_+_27131887 | 0.19 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr14_+_103243813 | 0.19 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr17_-_42277203 | 0.19 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr20_-_47804894 | 0.19 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr9_-_80263220 | 0.19 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MBD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 0.9 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.6 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.2 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.5 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.2 | 0.5 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 0.5 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.9 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 1.0 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.4 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 1.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.5 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.6 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.2 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.3 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.5 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.2 | GO:1903515 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) T-tubule organization(GO:0033292) calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.1 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.1 | 0.2 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.4 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 0.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.2 | GO:0061184 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 1.1 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.3 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 1.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.5 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.5 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0060922 | atrioventricular node development(GO:0003162) cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.7 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
| 0.0 | 1.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.5 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.2 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 1.1 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 1.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.7 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.5 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.0 | GO:0061355 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.0 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.5 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 0.6 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.2 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.4 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 1.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 1.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.2 | GO:0031775 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 1.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |