Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
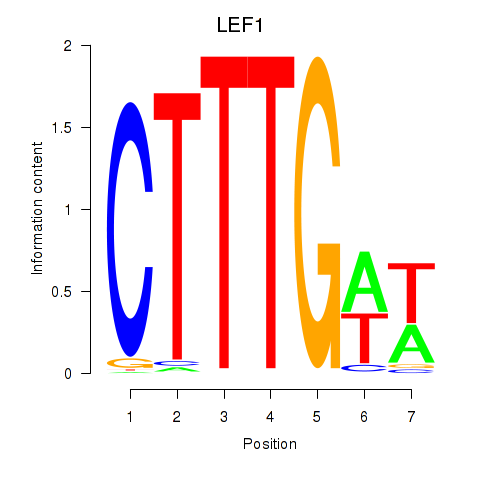
Results for LEF1
Z-value: 0.72
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | lymphoid enhancer binding factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg19_v2_chr4_-_109089573_109089585 | -0.53 | 4.7e-01 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_96869165 | 0.94 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr10_+_115312766 | 0.69 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr17_-_46035187 | 0.67 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr10_+_54074033 | 0.62 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr6_+_64282447 | 0.55 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr4_-_76649546 | 0.54 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr13_-_67802549 | 0.54 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr10_-_33405600 | 0.49 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chrX_+_80457442 | 0.48 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr16_+_53242350 | 0.45 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_+_95972822 | 0.43 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr17_+_4675175 | 0.42 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr2_+_181845763 | 0.41 |
ENST00000602499.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr10_+_115312825 | 0.41 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr21_+_17791648 | 0.38 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_+_90033968 | 0.34 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr15_-_37392086 | 0.34 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr3_+_107244229 | 0.33 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr5_-_34043310 | 0.33 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr5_+_138210919 | 0.33 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr3_-_112360116 | 0.33 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chrX_-_135962876 | 0.32 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr9_-_97356075 | 0.31 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chrX_-_45629661 | 0.31 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr8_-_95220775 | 0.31 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr5_+_31193847 | 0.31 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr1_-_65533390 | 0.30 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr14_+_58797974 | 0.30 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr1_+_27668505 | 0.29 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr12_+_94071129 | 0.29 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr5_+_137514834 | 0.28 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr20_+_36531499 | 0.28 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr14_+_38065052 | 0.28 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr6_+_43968306 | 0.27 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr5_+_137514687 | 0.27 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr6_-_29527702 | 0.27 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr14_-_67981916 | 0.27 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr1_-_93645818 | 0.27 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr1_+_6845578 | 0.26 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr11_-_111794446 | 0.26 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr4_+_57774042 | 0.26 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr2_+_70056762 | 0.26 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr1_-_156542328 | 0.26 |
ENST00000361170.2
|
IQGAP3
|
IQ motif containing GTPase activating protein 3 |
| chr4_-_140477928 | 0.26 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr14_-_65409502 | 0.25 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr15_-_37393406 | 0.25 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr7_+_16793160 | 0.25 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr3_-_167813672 | 0.25 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr1_+_47799542 | 0.24 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr1_-_219615984 | 0.24 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr1_+_9242221 | 0.24 |
ENST00000412639.2
|
RP3-510D11.2
|
RP3-510D11.2 |
| chr4_-_139163491 | 0.23 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr7_+_30185406 | 0.23 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr5_+_137673200 | 0.23 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr18_-_25616519 | 0.23 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr11_-_75921780 | 0.23 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr3_+_186648307 | 0.23 |
ENST00000457772.2
ENST00000455441.1 ENST00000427315.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr11_-_76155618 | 0.23 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr14_+_75746664 | 0.22 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr20_+_57875457 | 0.22 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr17_-_27503770 | 0.22 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr15_+_52311398 | 0.22 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr2_+_181845843 | 0.22 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chrX_-_135962923 | 0.22 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr2_-_107503558 | 0.22 |
ENST00000361686.4
ENST00000409087.3 |
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr12_+_94071341 | 0.22 |
ENST00000542893.2
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr20_+_36531544 | 0.22 |
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr1_+_53793885 | 0.22 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr1_-_108231101 | 0.22 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr15_-_37391614 | 0.21 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr16_-_15736881 | 0.21 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr1_-_54304212 | 0.21 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr20_+_37554955 | 0.21 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr6_+_64281906 | 0.21 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr15_-_52944231 | 0.21 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr18_+_9136758 | 0.21 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr14_-_38064198 | 0.21 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr16_-_70719925 | 0.21 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr5_+_172068232 | 0.21 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chrX_-_39956656 | 0.21 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr17_-_46623441 | 0.20 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr14_+_75746781 | 0.20 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr12_+_104359614 | 0.20 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr3_+_186648274 | 0.20 |
ENST00000169298.3
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr4_+_95128748 | 0.20 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr15_+_96873921 | 0.20 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr21_+_35552978 | 0.20 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr5_+_137514403 | 0.20 |
ENST00000513276.1
|
KIF20A
|
kinesin family member 20A |
| chrX_-_77041685 | 0.20 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr5_+_145317356 | 0.20 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr4_-_23891693 | 0.20 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr13_+_37581115 | 0.19 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr3_-_167813132 | 0.19 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr1_+_196621002 | 0.19 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr22_+_37959647 | 0.19 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr11_-_75017734 | 0.19 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr16_+_67063142 | 0.19 |
ENST00000412916.2
|
CBFB
|
core-binding factor, beta subunit |
| chr1_+_82266053 | 0.19 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr1_-_235491462 | 0.19 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr1_-_226496772 | 0.18 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr7_-_84121858 | 0.18 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_-_141747459 | 0.18 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_+_62432777 | 0.18 |
ENST00000532971.1
|
METTL12
|
methyltransferase like 12 |
| chr18_+_72922710 | 0.18 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr6_-_153304148 | 0.18 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chrX_+_56590002 | 0.18 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr21_+_35553045 | 0.18 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr3_-_125655882 | 0.18 |
ENST00000340333.3
|
ALG1L
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase-like |
| chr12_-_53994805 | 0.18 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr17_-_39165366 | 0.18 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr8_-_21988558 | 0.17 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chrX_+_38420783 | 0.17 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr14_-_67981870 | 0.17 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr20_+_11898507 | 0.17 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr15_+_36887069 | 0.17 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr3_+_189349162 | 0.17 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr1_+_93913713 | 0.17 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr2_-_211341411 | 0.17 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr15_+_57511609 | 0.17 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr16_-_30798492 | 0.17 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr6_-_153304697 | 0.17 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr2_-_170681324 | 0.17 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chr9_+_124088860 | 0.16 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr4_+_76649753 | 0.16 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr14_-_31676674 | 0.16 |
ENST00000399332.1
ENST00000556224.1 |
HECTD1
|
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr16_-_4987065 | 0.16 |
ENST00000590782.2
ENST00000345988.2 |
PPL
|
periplakin |
| chr17_+_57297807 | 0.16 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr17_-_70053866 | 0.16 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr8_-_17579726 | 0.16 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr17_+_29815013 | 0.16 |
ENST00000394744.2
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr11_-_102323489 | 0.16 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr1_+_196621156 | 0.16 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr5_+_162930114 | 0.16 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr5_+_92919043 | 0.16 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr22_+_27053422 | 0.16 |
ENST00000413665.1
ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr17_-_46682321 | 0.16 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr14_-_23451467 | 0.16 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr14_+_54976546 | 0.15 |
ENST00000216420.7
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr2_+_18059906 | 0.15 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr4_+_76649797 | 0.15 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr2_-_157189180 | 0.15 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_+_100435315 | 0.15 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr3_+_149535022 | 0.15 |
ENST00000466795.1
|
RNF13
|
ring finger protein 13 |
| chr2_-_48132814 | 0.15 |
ENST00000316377.4
ENST00000378314.3 |
FBXO11
|
F-box protein 11 |
| chr10_+_123970670 | 0.15 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_95128996 | 0.15 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr18_+_19668021 | 0.15 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr9_+_116638562 | 0.15 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr6_+_83777374 | 0.15 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr8_+_22019168 | 0.15 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr5_+_65018017 | 0.15 |
ENST00000380985.5
ENST00000502464.1 |
NLN
|
neurolysin (metallopeptidase M3 family) |
| chr18_-_53089723 | 0.15 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr10_-_32345305 | 0.15 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr5_-_135701164 | 0.15 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr8_+_42873548 | 0.14 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr11_-_102323740 | 0.14 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr12_+_104359576 | 0.14 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr1_+_32479430 | 0.14 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr1_+_93645314 | 0.14 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr3_+_152017924 | 0.14 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr12_-_112443830 | 0.14 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr21_+_17791838 | 0.14 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_-_63556414 | 0.14 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr19_+_1249869 | 0.14 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr5_+_95066823 | 0.14 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr2_-_86564776 | 0.14 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr6_+_27925019 | 0.14 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr15_+_93443419 | 0.14 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr21_-_34144157 | 0.14 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr2_-_26569611 | 0.14 |
ENST00000541401.1
ENST00000433584.1 ENST00000333478.6 |
GPR113
|
G protein-coupled receptor 113 |
| chr14_+_64971292 | 0.14 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr18_-_45456693 | 0.13 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr4_-_157892055 | 0.13 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr2_-_170681375 | 0.13 |
ENST00000410097.1
ENST00000308099.3 ENST00000409837.1 ENST00000538491.1 ENST00000260953.5 ENST00000409965.1 ENST00000392640.2 |
METTL5
|
methyltransferase like 5 |
| chr3_-_148804275 | 0.13 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr3_-_47823298 | 0.13 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr20_+_57875758 | 0.13 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr15_+_78730531 | 0.13 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr14_+_56078695 | 0.13 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr11_-_117747434 | 0.13 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr16_+_67063036 | 0.13 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr18_-_24237339 | 0.13 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr14_-_23451845 | 0.13 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr1_+_100435535 | 0.13 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr3_-_56809685 | 0.13 |
ENST00000413728.2
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr1_+_166808667 | 0.13 |
ENST00000537173.1
ENST00000536514.1 ENST00000449930.1 |
POGK
|
pogo transposable element with KRAB domain |
| chr4_+_95129061 | 0.13 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_78078304 | 0.13 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr2_-_47572105 | 0.13 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr1_+_84630053 | 0.13 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_141106643 | 0.12 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr13_+_39612442 | 0.12 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr2_+_145425573 | 0.12 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_+_93861993 | 0.12 |
ENST00000227638.3
ENST00000436171.2 |
PANX1
|
pannexin 1 |
| chr5_+_175288631 | 0.12 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr22_-_23484246 | 0.12 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr5_+_65222438 | 0.12 |
ENST00000380938.2
|
ERBB2IP
|
erbb2 interacting protein |
| chrX_-_63425561 | 0.12 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr20_-_656437 | 0.12 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chr7_-_102985035 | 0.12 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.6 | GO:0100012 | negative regulation of mesodermal cell fate specification(GO:0042662) motor learning(GO:0061743) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.1 | 0.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.3 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.3 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.4 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.1 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.1 | 0.2 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.2 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:1903217 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.0 | 0.3 | GO:0071638 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.2 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.8 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0003163 | sinoatrial node development(GO:0003163) sinoatrial node cell differentiation(GO:0060921) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.0 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.0 | GO:0031905 | early endosome lumen(GO:0031905) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.3 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.3 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.3 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |