Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
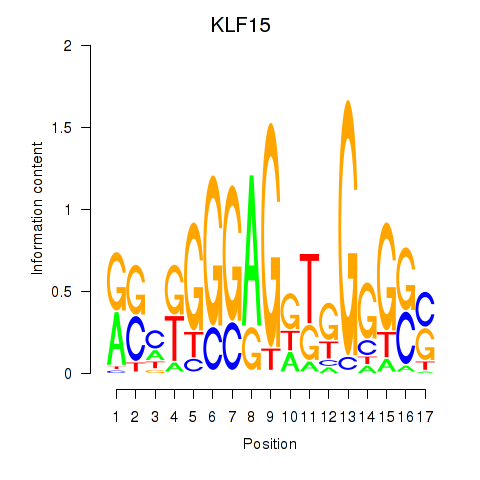
Results for KLF15
Z-value: 0.58
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | Kruppel like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF15 | hg19_v2_chr3_-_126076264_126076305 | 0.47 | 5.3e-01 | Click! |
Activity profile of KLF15 motif
Sorted Z-values of KLF15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_5832799 | 0.35 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr5_+_17217669 | 0.34 |
ENST00000322611.3
|
BASP1
|
brain abundant, membrane attached signal protein 1 |
| chr19_-_10047219 | 0.33 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr2_+_155554797 | 0.32 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr17_+_45771420 | 0.29 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr16_+_81478775 | 0.27 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr14_+_93673574 | 0.26 |
ENST00000554232.1
ENST00000556871.1 ENST00000555113.1 |
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr19_-_49362621 | 0.23 |
ENST00000594195.1
ENST00000595867.1 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr10_+_8096769 | 0.23 |
ENST00000346208.3
|
GATA3
|
GATA binding protein 3 |
| chrX_+_53078465 | 0.22 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr22_-_50970506 | 0.21 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr16_+_2802623 | 0.20 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr5_+_61602236 | 0.20 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr14_-_55738788 | 0.20 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr22_-_20792089 | 0.19 |
ENST00000405555.3
ENST00000266214.5 |
SCARF2
|
scavenger receptor class F, member 2 |
| chr11_-_64410787 | 0.19 |
ENST00000301894.2
|
NRXN2
|
neurexin 2 |
| chr17_-_36760865 | 0.19 |
ENST00000584266.1
|
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr18_-_51750948 | 0.19 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr10_-_79397547 | 0.19 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_30907862 | 0.18 |
ENST00000541765.1
ENST00000537108.1 |
CAPRIN2
|
caprin family member 2 |
| chr1_+_198126209 | 0.17 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr19_-_47975143 | 0.17 |
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_-_117267717 | 0.17 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr11_+_74951948 | 0.16 |
ENST00000562197.2
|
TPBGL
|
trophoblast glycoprotein-like |
| chr17_-_78450398 | 0.16 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr10_-_22292675 | 0.16 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr12_+_57854274 | 0.16 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr2_-_230786679 | 0.16 |
ENST00000543084.1
ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr2_-_200322723 | 0.15 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr11_-_31839488 | 0.15 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chrX_-_48693955 | 0.15 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr22_+_19118321 | 0.15 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr6_+_32811885 | 0.14 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_-_69445793 | 0.14 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chr10_+_28822236 | 0.14 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr10_-_101380121 | 0.14 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr5_+_131593364 | 0.14 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr11_-_65325664 | 0.14 |
ENST00000301873.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr4_+_39640787 | 0.14 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr19_-_46272106 | 0.13 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr19_-_45661995 | 0.13 |
ENST00000438936.2
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr17_-_4545170 | 0.13 |
ENST00000576394.1
ENST00000574640.1 |
ALOX15
|
arachidonate 15-lipoxygenase |
| chr1_+_206680879 | 0.13 |
ENST00000355294.4
ENST00000367117.3 |
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr22_+_42470289 | 0.13 |
ENST00000419475.1
|
FAM109B
|
family with sequence similarity 109, member B |
| chr11_-_134095335 | 0.13 |
ENST00000534227.1
ENST00000532445.1 |
NCAPD3
|
non-SMC condensin II complex, subunit D3 |
| chr2_+_45878790 | 0.13 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr8_+_81397876 | 0.12 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr10_+_28822417 | 0.12 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chrX_-_19905703 | 0.12 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr3_-_88108212 | 0.12 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr7_-_151107106 | 0.12 |
ENST00000334493.6
|
WDR86
|
WD repeat domain 86 |
| chr22_-_50699972 | 0.12 |
ENST00000395778.3
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr15_+_90728145 | 0.12 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr12_+_52404270 | 0.12 |
ENST00000552049.1
ENST00000546756.1 |
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr21_-_27107344 | 0.11 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr2_-_230786378 | 0.11 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr4_+_74702214 | 0.11 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chrX_+_154114635 | 0.11 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr2_-_198175495 | 0.11 |
ENST00000409153.1
ENST00000409919.1 ENST00000539527.1 |
ANKRD44
|
ankyrin repeat domain 44 |
| chr8_+_21777243 | 0.11 |
ENST00000521303.1
|
XPO7
|
exportin 7 |
| chr12_+_109592477 | 0.11 |
ENST00000544726.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr19_-_46272462 | 0.11 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr2_+_232573208 | 0.11 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr19_-_47975417 | 0.11 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr19_-_14201507 | 0.11 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr19_-_38806540 | 0.11 |
ENST00000592694.1
|
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr18_-_21977748 | 0.10 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr17_-_4463856 | 0.10 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr1_-_179198702 | 0.10 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr16_+_30709530 | 0.10 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr1_-_244013384 | 0.10 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr22_+_39745930 | 0.10 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr7_+_128312346 | 0.10 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr21_-_27107198 | 0.10 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr1_-_243418650 | 0.10 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr19_+_852291 | 0.10 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr17_+_42634844 | 0.10 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr7_-_100493482 | 0.10 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr7_-_154863264 | 0.10 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr17_-_17399701 | 0.10 |
ENST00000225688.3
ENST00000579152.1 |
RASD1
|
RAS, dexamethasone-induced 1 |
| chr3_-_49726104 | 0.10 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr13_-_52027134 | 0.09 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr19_-_38806560 | 0.09 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr7_+_92158083 | 0.09 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr20_+_44650348 | 0.09 |
ENST00000454036.2
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr11_-_46142505 | 0.09 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr1_-_110933663 | 0.09 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr11_-_46142615 | 0.09 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr15_-_65067773 | 0.09 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
| chr17_+_78234625 | 0.09 |
ENST00000508628.2
ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr7_-_92157760 | 0.09 |
ENST00000248633.4
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr2_+_74212073 | 0.09 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr1_+_37940153 | 0.09 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr21_+_30671189 | 0.09 |
ENST00000286800.3
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr2_-_230786619 | 0.09 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr19_-_40790993 | 0.09 |
ENST00000596634.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_+_46926326 | 0.09 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_-_233792837 | 0.09 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr19_+_8117636 | 0.09 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr19_-_4124079 | 0.09 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr9_+_88556444 | 0.09 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr2_+_39893043 | 0.08 |
ENST00000281961.2
|
TMEM178A
|
transmembrane protein 178A |
| chr10_+_8096631 | 0.08 |
ENST00000379328.3
|
GATA3
|
GATA binding protein 3 |
| chr16_-_755726 | 0.08 |
ENST00000324361.5
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_150898733 | 0.08 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chr4_+_56719782 | 0.08 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr4_-_1657135 | 0.08 |
ENST00000489029.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr2_+_198669365 | 0.08 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr20_-_62680984 | 0.08 |
ENST00000340356.7
|
SOX18
|
SRY (sex determining region Y)-box 18 |
| chr11_-_62457371 | 0.08 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr1_+_33283043 | 0.08 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr9_-_139891165 | 0.08 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr7_+_89841024 | 0.08 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr1_-_110933611 | 0.08 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr10_+_76871454 | 0.08 |
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr19_-_6424783 | 0.08 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr12_+_120933859 | 0.08 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr7_+_144052381 | 0.08 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr10_+_76871353 | 0.08 |
ENST00000542569.1
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr7_-_92157747 | 0.08 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr2_+_201171268 | 0.08 |
ENST00000423749.1
ENST00000428692.1 ENST00000457757.1 ENST00000453663.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_+_3449695 | 0.08 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr15_+_40544749 | 0.08 |
ENST00000559617.1
ENST00000560684.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr7_+_116139744 | 0.08 |
ENST00000343213.2
|
CAV2
|
caveolin 2 |
| chr10_-_121356518 | 0.08 |
ENST00000369092.4
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr13_-_52026730 | 0.08 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr8_-_105601134 | 0.07 |
ENST00000276654.5
ENST00000424843.2 |
LRP12
|
low density lipoprotein receptor-related protein 12 |
| chr14_+_92980111 | 0.07 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr2_-_45166338 | 0.07 |
ENST00000437916.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr19_-_11689752 | 0.07 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_140770183 | 0.07 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr7_+_29519662 | 0.07 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr21_-_27107283 | 0.07 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr13_-_108867846 | 0.07 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr6_-_29595779 | 0.07 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_-_143059780 | 0.07 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr21_-_27107881 | 0.07 |
ENST00000400090.3
ENST00000400087.3 ENST00000400093.3 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr13_-_45151259 | 0.07 |
ENST00000493016.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr19_-_3062465 | 0.07 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr2_-_39347524 | 0.07 |
ENST00000395038.2
ENST00000402219.2 |
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr2_-_9143786 | 0.07 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr12_+_120933904 | 0.07 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr11_-_65641044 | 0.07 |
ENST00000527378.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr16_+_3162557 | 0.07 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr12_+_53774423 | 0.07 |
ENST00000426431.2
|
SP1
|
Sp1 transcription factor |
| chr3_-_185826286 | 0.07 |
ENST00000537818.1
ENST00000422039.1 ENST00000434744.1 |
ETV5
|
ets variant 5 |
| chr19_-_47249679 | 0.07 |
ENST00000263280.6
|
STRN4
|
striatin, calmodulin binding protein 4 |
| chr11_+_50257784 | 0.07 |
ENST00000529219.1
|
RP11-347H15.4
|
RP11-347H15.4 |
| chr6_-_94129244 | 0.07 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chrX_+_119495934 | 0.07 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr5_-_131132658 | 0.07 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr2_+_232573222 | 0.06 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr7_-_151217001 | 0.06 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr2_-_208030647 | 0.06 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_+_24837226 | 0.06 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr14_-_51562037 | 0.06 |
ENST00000338969.5
|
TRIM9
|
tripartite motif containing 9 |
| chr9_+_137533615 | 0.06 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr19_-_55866061 | 0.06 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr16_-_30107491 | 0.06 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr17_-_27332931 | 0.06 |
ENST00000442608.3
ENST00000335960.6 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr12_-_50101003 | 0.06 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr1_-_21503337 | 0.06 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr16_+_30969055 | 0.06 |
ENST00000452917.1
|
SETD1A
|
SET domain containing 1A |
| chr21_-_40685504 | 0.06 |
ENST00000380800.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr13_+_98795434 | 0.06 |
ENST00000376586.2
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr11_+_119076745 | 0.06 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr3_-_27763803 | 0.06 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr16_+_50300427 | 0.06 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr17_-_76921459 | 0.06 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr7_+_89841000 | 0.06 |
ENST00000287908.3
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr2_-_153573965 | 0.06 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr14_-_73493784 | 0.06 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr19_+_41082755 | 0.06 |
ENST00000291842.5
ENST00000600733.1 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr2_-_73511559 | 0.06 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr19_-_47975106 | 0.06 |
ENST00000539381.1
ENST00000594353.1 ENST00000542837.1 |
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr8_+_9413410 | 0.06 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr17_-_58469687 | 0.06 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr10_+_114709999 | 0.05 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_70780770 | 0.05 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr19_+_589893 | 0.05 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr11_-_117667806 | 0.05 |
ENST00000527706.1
ENST00000321322.6 |
DSCAML1
|
Down syndrome cell adhesion molecule like 1 |
| chr17_-_4852332 | 0.05 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr12_-_131323719 | 0.05 |
ENST00000392373.2
|
STX2
|
syntaxin 2 |
| chr19_-_50400212 | 0.05 |
ENST00000391826.2
|
IL4I1
|
interleukin 4 induced 1 |
| chr1_+_180199393 | 0.05 |
ENST00000263726.2
|
LHX4
|
LIM homeobox 4 |
| chr15_+_72766651 | 0.05 |
ENST00000379887.4
|
ARIH1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr1_+_203765437 | 0.05 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr17_-_42277203 | 0.05 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr2_-_198650037 | 0.05 |
ENST00000392296.4
|
BOLL
|
boule-like RNA-binding protein |
| chr11_-_72353451 | 0.05 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr3_+_101546827 | 0.05 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr19_-_42636617 | 0.05 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr18_-_57364588 | 0.05 |
ENST00000439986.4
|
CCBE1
|
collagen and calcium binding EGF domains 1 |
| chr17_-_27278445 | 0.05 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr1_-_27930102 | 0.05 |
ENST00000247087.5
ENST00000374011.2 |
AHDC1
|
AT hook, DNA binding motif, containing 1 |
| chr2_-_37193606 | 0.05 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr1_-_151689259 | 0.05 |
ENST00000420342.1
ENST00000290583.4 |
CELF3
|
CUGBP, Elav-like family member 3 |
| chr18_+_48405419 | 0.05 |
ENST00000321341.5
|
ME2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr3_-_149688502 | 0.05 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr19_+_45754505 | 0.05 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.3 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.3 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.2 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.3 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:2000625 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.2 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:1902462 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0070945 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.0 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.0 | GO:0044727 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.0 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) positive regulation of dopamine receptor signaling pathway(GO:0060161) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.0 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.0 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.0 | 0.0 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.1 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0050473 | hepoxilin-epoxide hydrolase activity(GO:0047977) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |