Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for KLF14_SP8
Z-value: 0.85
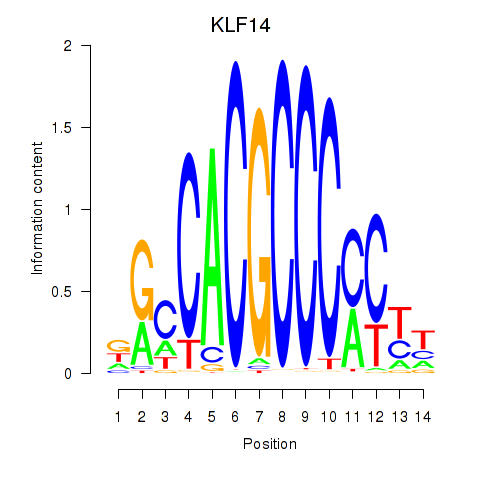
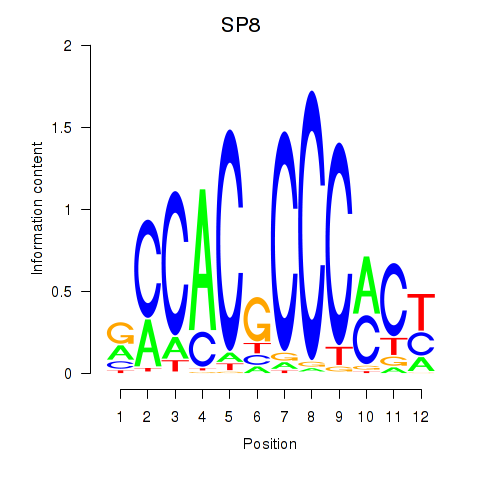
Transcription factors associated with KLF14_SP8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF14
|
ENSG00000174595.4 | Kruppel like factor 14 |
|
SP8
|
ENSG00000164651.12 | Sp8 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP8 | hg19_v2_chr7_-_20826504_20826526 | 0.76 | 2.4e-01 | Click! |
| KLF14 | hg19_v2_chr7_-_130418888_130418888 | 0.75 | 2.5e-01 | Click! |
Activity profile of KLF14_SP8 motif
Sorted Z-values of KLF14_SP8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_228362251 | 0.66 |
ENST00000546123.1
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr19_+_50433476 | 0.46 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr17_-_882966 | 0.40 |
ENST00000336868.3
|
NXN
|
nucleoredoxin |
| chr19_-_10946871 | 0.38 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr19_+_39786962 | 0.38 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr5_+_89770664 | 0.30 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr2_-_110371664 | 0.30 |
ENST00000545389.1
ENST00000423520.1 |
SEPT10
|
septin 10 |
| chr19_+_45349630 | 0.29 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr5_+_158527485 | 0.29 |
ENST00000517335.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr18_+_61144160 | 0.28 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr13_+_52158610 | 0.27 |
ENST00000298125.5
|
WDFY2
|
WD repeat and FYVE domain containing 2 |
| chr9_+_19408919 | 0.27 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr19_-_49362621 | 0.27 |
ENST00000594195.1
ENST00000595867.1 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr9_-_35658007 | 0.26 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr17_+_7184986 | 0.26 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr15_-_32695396 | 0.26 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr11_+_10772847 | 0.25 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr12_-_56753858 | 0.24 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr7_+_100770328 | 0.24 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr1_+_45140400 | 0.24 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr22_+_27068766 | 0.23 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr12_+_56522001 | 0.23 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr1_-_200992827 | 0.23 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr19_-_36231437 | 0.23 |
ENST00000591748.1
|
IGFLR1
|
IGF-like family receptor 1 |
| chr10_+_74033672 | 0.22 |
ENST00000307365.3
|
DDIT4
|
DNA-damage-inducible transcript 4 |
| chr22_+_31477296 | 0.22 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr5_-_180237082 | 0.22 |
ENST00000506889.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr4_+_89378261 | 0.22 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chrX_+_54835493 | 0.22 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr2_+_64068116 | 0.21 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr19_+_45349432 | 0.21 |
ENST00000252485.4
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr22_+_30279144 | 0.20 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr6_-_163834852 | 0.20 |
ENST00000604200.1
|
CAHM
|
colon adenocarcinoma hypermethylated (non-protein coding) |
| chr3_+_66271410 | 0.20 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr14_+_54976603 | 0.20 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr21_+_44394742 | 0.20 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr18_-_12377001 | 0.20 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr12_-_25348007 | 0.19 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr12_+_7341759 | 0.19 |
ENST00000455147.2
ENST00000540398.1 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr22_-_30685596 | 0.19 |
ENST00000404953.3
ENST00000407689.3 |
GATSL3
|
GATS protein-like 3 |
| chr10_+_13142075 | 0.19 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr15_-_40401062 | 0.19 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr19_-_56879416 | 0.18 |
ENST00000591036.1
|
AC006116.21
|
AC006116.21 |
| chrX_+_68048803 | 0.18 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr10_+_75910960 | 0.18 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chr2_-_110371412 | 0.18 |
ENST00000415095.1
ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10
|
septin 10 |
| chr10_-_73975657 | 0.18 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr10_-_73976025 | 0.18 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr12_+_107349606 | 0.18 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr12_+_110718428 | 0.17 |
ENST00000552636.1
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr14_-_21562671 | 0.17 |
ENST00000554923.1
|
ZNF219
|
zinc finger protein 219 |
| chr1_-_161147275 | 0.17 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr19_+_54695098 | 0.17 |
ENST00000396388.2
|
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr5_+_109025067 | 0.16 |
ENST00000261483.4
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1 |
| chr19_+_19516561 | 0.16 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr12_-_56615485 | 0.16 |
ENST00000549038.1
ENST00000552244.1 |
RNF41
|
ring finger protein 41 |
| chr9_+_131038425 | 0.16 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr16_+_67280799 | 0.16 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chrX_+_68835911 | 0.16 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr15_-_28344439 | 0.15 |
ENST00000431101.1
ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2
|
oculocutaneous albinism II |
| chr19_+_5904866 | 0.15 |
ENST00000339485.3
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr12_-_118810688 | 0.15 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr11_-_77705687 | 0.15 |
ENST00000529807.1
ENST00000527522.1 ENST00000534064.1 |
INTS4
|
integrator complex subunit 4 |
| chr3_+_14444063 | 0.15 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr12_+_111843749 | 0.15 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr12_+_110718921 | 0.15 |
ENST00000308664.6
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr2_-_73340146 | 0.15 |
ENST00000258098.6
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I) |
| chr6_-_30523865 | 0.15 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr11_-_77531752 | 0.15 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr6_-_167369612 | 0.15 |
ENST00000507747.1
|
RP11-514O12.4
|
RP11-514O12.4 |
| chr11_+_65480222 | 0.15 |
ENST00000534681.1
|
KAT5
|
K(lysine) acetyltransferase 5 |
| chr3_+_10206545 | 0.15 |
ENST00000256458.4
|
IRAK2
|
interleukin-1 receptor-associated kinase 2 |
| chr7_+_111846741 | 0.14 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr12_+_51632638 | 0.14 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr15_+_72410660 | 0.14 |
ENST00000564082.1
|
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr14_+_96829814 | 0.14 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr6_+_19837592 | 0.14 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr12_+_7342271 | 0.14 |
ENST00000434354.2
ENST00000544456.1 ENST00000545574.1 ENST00000420616.2 |
PEX5
|
peroxisomal biogenesis factor 5 |
| chr12_+_122064398 | 0.14 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr19_+_54372877 | 0.14 |
ENST00000414489.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr7_+_98971863 | 0.14 |
ENST00000443222.1
ENST00000414376.1 |
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr22_+_27068704 | 0.14 |
ENST00000444388.1
ENST00000450963.1 ENST00000449017.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr19_+_54694119 | 0.14 |
ENST00000456872.1
ENST00000302937.4 ENST00000429671.2 |
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr9_+_34652164 | 0.13 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr10_-_135171178 | 0.13 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr17_+_41177220 | 0.13 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr5_-_176924562 | 0.13 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr5_+_154238149 | 0.13 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr10_-_7708918 | 0.13 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr1_+_172502336 | 0.13 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr15_+_89631647 | 0.13 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr17_+_48133330 | 0.13 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr11_-_102962929 | 0.13 |
ENST00000260247.5
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr17_+_37821593 | 0.13 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr8_+_124428959 | 0.13 |
ENST00000287387.2
ENST00000523984.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr2_+_220071490 | 0.13 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr1_-_42921915 | 0.13 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr9_-_38069208 | 0.13 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr17_-_8027402 | 0.13 |
ENST00000541682.2
ENST00000317814.4 ENST00000577735.1 |
HES7
|
hes family bHLH transcription factor 7 |
| chr17_-_42276574 | 0.13 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr15_+_41099919 | 0.12 |
ENST00000561617.1
|
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr3_-_171178157 | 0.12 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr7_-_33102399 | 0.12 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr12_+_51632666 | 0.12 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr19_-_46000251 | 0.12 |
ENST00000590526.1
ENST00000344680.4 ENST00000245923.4 |
RTN2
|
reticulon 2 |
| chr3_+_66271489 | 0.12 |
ENST00000536651.1
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr2_+_70142232 | 0.12 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr11_-_62521614 | 0.12 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr19_-_47164386 | 0.12 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr12_+_6833323 | 0.12 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr4_-_10117949 | 0.12 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr3_-_194991876 | 0.12 |
ENST00000310380.6
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chrX_-_48814810 | 0.12 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr12_+_3186521 | 0.12 |
ENST00000537971.1
ENST00000011898.5 |
TSPAN9
|
tetraspanin 9 |
| chr1_-_155880672 | 0.12 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr16_+_15737124 | 0.12 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr19_-_54693521 | 0.12 |
ENST00000391754.1
ENST00000245615.1 ENST00000431666.2 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr1_+_112938803 | 0.12 |
ENST00000271277.6
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr5_-_149792295 | 0.12 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr11_+_63606558 | 0.12 |
ENST00000350490.7
ENST00000502399.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chrX_-_152989798 | 0.12 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr3_-_48594248 | 0.12 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr19_-_42759266 | 0.12 |
ENST00000594664.1
|
AC006486.9
|
Uncharacterized protein |
| chr17_+_4402133 | 0.12 |
ENST00000329078.3
|
SPNS2
|
spinster homolog 2 (Drosophila) |
| chr19_-_52598958 | 0.12 |
ENST00000594440.1
ENST00000426391.2 ENST00000389534.4 |
ZNF841
|
zinc finger protein 841 |
| chr15_+_63414760 | 0.12 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr19_-_54693146 | 0.12 |
ENST00000414665.1
ENST00000453320.1 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr11_-_19263145 | 0.12 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr17_-_6915616 | 0.12 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr5_-_43313269 | 0.11 |
ENST00000511774.1
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr4_-_4291761 | 0.11 |
ENST00000513174.1
|
LYAR
|
Ly1 antibody reactive |
| chr11_+_93517516 | 0.11 |
ENST00000533359.1
|
MED17
|
mediator complex subunit 17 |
| chr16_-_54962625 | 0.11 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr3_+_197464046 | 0.11 |
ENST00000428738.1
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr19_+_39138320 | 0.11 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr1_+_155051379 | 0.11 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr19_+_10197463 | 0.11 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr8_+_145202939 | 0.11 |
ENST00000423230.2
ENST00000398656.4 |
MROH1
|
maestro heat-like repeat family member 1 |
| chr16_+_31044413 | 0.11 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr4_+_17579110 | 0.11 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr1_+_32538492 | 0.11 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr22_-_30783075 | 0.11 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr5_-_81046841 | 0.11 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr11_-_71752571 | 0.11 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr1_+_222886694 | 0.11 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr6_-_31782813 | 0.11 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr7_-_33102338 | 0.11 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr11_+_71640112 | 0.11 |
ENST00000530137.1
|
RNF121
|
ring finger protein 121 |
| chr6_+_24126350 | 0.11 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr11_+_63606477 | 0.10 |
ENST00000508192.1
ENST00000361128.5 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr2_+_239335636 | 0.10 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr1_+_6673745 | 0.10 |
ENST00000377648.4
|
PHF13
|
PHD finger protein 13 |
| chr3_-_50374869 | 0.10 |
ENST00000327761.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr21_+_48055527 | 0.10 |
ENST00000397638.2
ENST00000458387.2 ENST00000451211.2 ENST00000291705.6 ENST00000397637.1 ENST00000334494.4 ENST00000397628.1 ENST00000440086.1 |
PRMT2
|
protein arginine methyltransferase 2 |
| chr18_-_43684230 | 0.10 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr19_-_10946949 | 0.10 |
ENST00000214869.2
ENST00000591695.1 |
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr3_+_97483366 | 0.10 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr20_-_47894569 | 0.10 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr1_+_112939121 | 0.10 |
ENST00000441739.1
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr16_-_84178728 | 0.10 |
ENST00000562224.1
ENST00000434463.3 ENST00000564998.1 ENST00000219439.4 |
HSDL1
|
hydroxysteroid dehydrogenase like 1 |
| chr22_+_46476192 | 0.10 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr1_+_169337412 | 0.10 |
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr2_-_68290106 | 0.10 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr14_-_31889737 | 0.10 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr4_+_39046615 | 0.10 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr1_+_33352036 | 0.10 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chrX_+_54834791 | 0.10 |
ENST00000218439.4
ENST00000375058.1 ENST00000375060.1 |
MAGED2
|
melanoma antigen family D, 2 |
| chr5_+_61601965 | 0.10 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr19_+_50353944 | 0.10 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr12_-_54813229 | 0.10 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr6_-_31138439 | 0.10 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr6_-_110500826 | 0.10 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr18_+_9334786 | 0.10 |
ENST00000581641.1
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr15_+_41099254 | 0.10 |
ENST00000570108.1
ENST00000564258.1 ENST00000355341.4 ENST00000336455.5 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr1_+_172502244 | 0.10 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr10_-_101380121 | 0.10 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr20_-_4804244 | 0.10 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr17_+_48133459 | 0.10 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr12_-_56615693 | 0.10 |
ENST00000394013.2
ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41
|
ring finger protein 41 |
| chr12_-_58240470 | 0.10 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr18_+_33877654 | 0.10 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr6_-_33266687 | 0.10 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr7_-_106301405 | 0.10 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr19_+_36606354 | 0.10 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chrX_+_48660287 | 0.10 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr1_+_200993071 | 0.10 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr7_-_91764108 | 0.10 |
ENST00000450723.1
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr7_+_1570322 | 0.10 |
ENST00000343242.4
|
MAFK
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr2_+_64068074 | 0.10 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr11_+_60699222 | 0.10 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chr12_+_69979210 | 0.10 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr7_+_128431444 | 0.10 |
ENST00000459946.1
ENST00000378685.4 ENST00000464832.1 ENST00000472049.1 ENST00000488925.1 |
CCDC136
|
coiled-coil domain containing 136 |
| chr15_+_31619013 | 0.10 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr4_-_166034004 | 0.09 |
ENST00000505095.1
|
TMEM192
|
transmembrane protein 192 |
| chr11_-_115375107 | 0.09 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chrX_-_48815633 | 0.09 |
ENST00000428668.2
|
OTUD5
|
OTU domain containing 5 |
| chr20_-_44539538 | 0.09 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr12_+_48499883 | 0.09 |
ENST00000546755.1
ENST00000549366.1 ENST00000552792.1 |
PFKM
|
phosphofructokinase, muscle |
| chr11_-_94964354 | 0.09 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chr20_-_17662705 | 0.09 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF14_SP8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum(GO:1903515) |
| 0.1 | 0.4 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.4 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.4 | GO:1901094 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.1 | 0.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 0.2 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.2 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.2 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.4 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.2 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.3 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0050757 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.0 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.4 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.0 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.2 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.2 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.0 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.0 | 0.0 | GO:0090191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.0 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0046717 | acid secretion(GO:0046717) |
| 0.0 | 0.1 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.3 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.5 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0086039 | lutropin-choriogonadotropic hormone receptor binding(GO:0031775) calcium-transporting ATPase activity involved in regulation of cardiac muscle cell membrane potential(GO:0086039) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.3 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.1 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.0 | 0.1 | GO:0070704 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.0 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.0 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |