Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
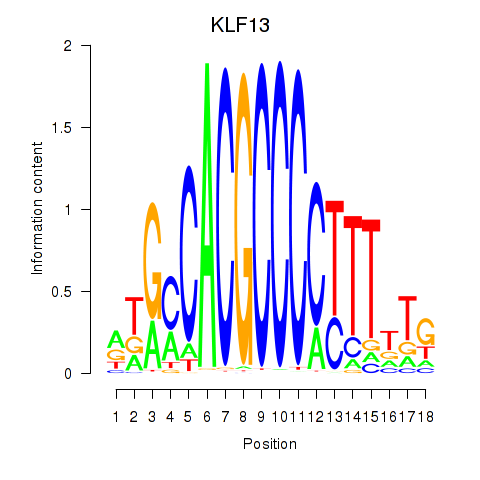
Results for KLF13
Z-value: 0.84
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | Kruppel like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF13 | hg19_v2_chr15_+_31619013_31619095 | 0.46 | 5.4e-01 | Click! |
Activity profile of KLF13 motif
Sorted Z-values of KLF13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_31697977 | 0.60 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_+_74551650 | 0.51 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr7_+_2281843 | 0.46 |
ENST00000356714.1
ENST00000397049.1 |
NUDT1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr6_-_31697563 | 0.44 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_44587141 | 0.42 |
ENST00000227155.4
ENST00000342935.3 ENST00000532544.1 |
CD82
|
CD82 molecule |
| chr3_-_138048631 | 0.38 |
ENST00000484930.1
ENST00000475751.1 |
NME9
|
NME/NM23 family member 9 |
| chr19_-_15236562 | 0.36 |
ENST00000263383.3
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr15_-_65117807 | 0.35 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr6_+_31126291 | 0.35 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr16_-_75284758 | 0.32 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr1_-_42921915 | 0.32 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr15_+_30896329 | 0.30 |
ENST00000566740.1
|
GOLGA8H
|
golgin A8 family, member H |
| chr12_-_49581152 | 0.29 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr12_-_124457163 | 0.29 |
ENST00000535556.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr9_+_131038425 | 0.29 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr19_-_15236470 | 0.27 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr22_-_41985865 | 0.27 |
ENST00000216259.7
|
PMM1
|
phosphomannomutase 1 |
| chr11_+_73498973 | 0.27 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr10_+_115803650 | 0.26 |
ENST00000369295.2
|
ADRB1
|
adrenoceptor beta 1 |
| chr2_-_128615517 | 0.26 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr2_-_128615681 | 0.26 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr6_-_31125850 | 0.26 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr15_+_40453204 | 0.25 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr5_+_111496631 | 0.24 |
ENST00000508590.1
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chr21_+_45905448 | 0.24 |
ENST00000449713.1
|
AP001065.15
|
AP001065.15 |
| chr11_-_6426635 | 0.22 |
ENST00000608645.1
ENST00000608394.1 ENST00000529519.1 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr19_+_12917364 | 0.22 |
ENST00000221486.4
|
RNASEH2A
|
ribonuclease H2, subunit A |
| chr4_+_1795012 | 0.22 |
ENST00000481110.2
ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3
|
fibroblast growth factor receptor 3 |
| chr20_-_8000426 | 0.22 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr7_-_23053693 | 0.21 |
ENST00000409763.1
ENST00000409923.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr13_-_30424821 | 0.21 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr10_-_13344341 | 0.21 |
ENST00000396920.3
|
PHYH
|
phytanoyl-CoA 2-hydroxylase |
| chr19_-_50311896 | 0.21 |
ENST00000529634.2
|
FUZ
|
fuzzy planar cell polarity protein |
| chr19_+_45843994 | 0.21 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr9_+_132597722 | 0.20 |
ENST00000372429.3
ENST00000315480.4 ENST00000358355.1 |
USP20
|
ubiquitin specific peptidase 20 |
| chr11_-_27494309 | 0.20 |
ENST00000389858.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr11_-_27494279 | 0.20 |
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr11_-_33722286 | 0.19 |
ENST00000451594.2
ENST00000379011.4 |
C11orf91
|
chromosome 11 open reading frame 91 |
| chr12_+_58013693 | 0.19 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr6_+_33172407 | 0.19 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr7_+_111846741 | 0.19 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr7_-_91763822 | 0.18 |
ENST00000003100.8
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr15_+_41056255 | 0.18 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr19_+_36249044 | 0.18 |
ENST00000444637.2
ENST00000396908.4 ENST00000544099.1 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr1_+_874649 | 0.17 |
ENST00000455979.1
|
SAMD11
|
sterile alpha motif domain containing 11 |
| chr10_+_134351319 | 0.17 |
ENST00000368594.3
ENST00000368593.3 |
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa |
| chr14_+_54976546 | 0.17 |
ENST00000216420.7
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr11_+_44587206 | 0.17 |
ENST00000525210.1
ENST00000527737.1 ENST00000524704.1 |
CD82
|
CD82 molecule |
| chr9_-_77567743 | 0.17 |
ENST00000376854.5
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr11_+_65480222 | 0.17 |
ENST00000534681.1
|
KAT5
|
K(lysine) acetyltransferase 5 |
| chr5_+_148206156 | 0.16 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr3_+_66271410 | 0.16 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr12_-_49961883 | 0.16 |
ENST00000553173.1
ENST00000550165.1 ENST00000546244.1 ENST00000343810.4 |
MCRS1
|
microspherule protein 1 |
| chr5_+_154238149 | 0.16 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr19_+_36249057 | 0.16 |
ENST00000301165.5
ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr11_+_65383227 | 0.16 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr7_-_139168434 | 0.15 |
ENST00000340940.4
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr1_+_45805342 | 0.15 |
ENST00000372090.5
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr9_-_131592058 | 0.15 |
ENST00000361256.5
|
C9orf114
|
chromosome 9 open reading frame 114 |
| chr14_-_23299009 | 0.15 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_-_74551096 | 0.15 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr19_+_45844018 | 0.15 |
ENST00000585434.1
|
KLC3
|
kinesin light chain 3 |
| chr20_+_37590942 | 0.15 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr7_+_99775366 | 0.15 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chr16_+_84853580 | 0.15 |
ENST00000262424.5
ENST00000566151.1 ENST00000567845.1 ENST00000564567.1 ENST00000569090.1 |
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr9_+_19408999 | 0.14 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr16_-_28936493 | 0.14 |
ENST00000544477.1
ENST00000357573.6 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr15_-_82939157 | 0.14 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr3_-_138048682 | 0.14 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr16_+_4526341 | 0.14 |
ENST00000458134.3
ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr9_+_91926103 | 0.13 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr17_-_77770830 | 0.13 |
ENST00000269385.4
|
CBX8
|
chromobox homolog 8 |
| chr14_-_67981916 | 0.13 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chr17_-_17109579 | 0.13 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chrX_+_47077632 | 0.12 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr17_+_55162453 | 0.12 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr9_-_123964114 | 0.12 |
ENST00000373840.4
|
RAB14
|
RAB14, member RAS oncogene family |
| chr17_+_74734052 | 0.12 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr15_+_41851211 | 0.12 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr15_-_82641706 | 0.12 |
ENST00000439287.4
|
GOLGA6L10
|
golgin A6 family-like 10 |
| chr2_-_8977714 | 0.12 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr19_+_7985198 | 0.11 |
ENST00000221573.6
ENST00000595637.1 |
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr3_+_50606577 | 0.11 |
ENST00000434410.1
ENST00000232854.4 |
HEMK1
|
HemK methyltransferase family member 1 |
| chr19_-_46142362 | 0.11 |
ENST00000586770.1
ENST00000591721.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr21_+_48055527 | 0.11 |
ENST00000397638.2
ENST00000458387.2 ENST00000451211.2 ENST00000291705.6 ENST00000397637.1 ENST00000334494.4 ENST00000397628.1 ENST00000440086.1 |
PRMT2
|
protein arginine methyltransferase 2 |
| chr19_+_8509842 | 0.11 |
ENST00000325495.4
ENST00000600092.1 ENST00000594907.1 ENST00000596984.1 ENST00000601645.1 |
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M |
| chr17_+_79679299 | 0.11 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr6_+_52535878 | 0.11 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr15_+_52311398 | 0.11 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr12_-_29534074 | 0.10 |
ENST00000546839.1
ENST00000360150.4 ENST00000552155.1 ENST00000550353.1 ENST00000548441.1 ENST00000552132.1 |
ERGIC2
|
ERGIC and golgi 2 |
| chr19_+_54695098 | 0.10 |
ENST00000396388.2
|
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr7_-_99698338 | 0.10 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr3_+_50606901 | 0.10 |
ENST00000455834.1
|
HEMK1
|
HemK methyltransferase family member 1 |
| chr2_+_239335636 | 0.10 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr17_+_75446819 | 0.10 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
| chr17_+_79679369 | 0.09 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr7_-_2281802 | 0.09 |
ENST00000242257.8
ENST00000440306.2 |
FTSJ2
|
FtsJ RNA methyltransferase homolog 2 (E. coli) |
| chr17_+_7184986 | 0.09 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr15_+_32885657 | 0.09 |
ENST00000448387.2
ENST00000569659.1 |
GOLGA8N
|
golgin A8 family, member N |
| chr20_+_34129770 | 0.09 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr3_+_152552685 | 0.09 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr12_+_49961990 | 0.09 |
ENST00000551063.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr6_-_31697255 | 0.09 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr20_-_44600810 | 0.09 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr3_+_66271489 | 0.09 |
ENST00000536651.1
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr2_-_97405775 | 0.09 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr4_+_57774042 | 0.09 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr5_-_32313019 | 0.09 |
ENST00000280285.5
ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr19_+_36632056 | 0.09 |
ENST00000586851.1
ENST00000590211.1 |
CAPNS1
|
calpain, small subunit 1 |
| chr6_+_28193037 | 0.09 |
ENST00000531981.1
ENST00000425468.2 ENST00000252207.5 ENST00000531979.1 ENST00000527436.1 |
ZSCAN9
|
zinc finger and SCAN domain containing 9 |
| chr1_-_161147275 | 0.09 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr2_+_200775971 | 0.09 |
ENST00000319974.5
|
C2orf69
|
chromosome 2 open reading frame 69 |
| chr9_-_132597529 | 0.09 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr21_-_33765303 | 0.09 |
ENST00000382751.3
|
URB1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr14_-_94970559 | 0.08 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr17_+_27055798 | 0.08 |
ENST00000268766.6
|
NEK8
|
NIMA-related kinase 8 |
| chr7_-_23053719 | 0.08 |
ENST00000432176.2
ENST00000440481.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr17_+_42264556 | 0.08 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_-_224517823 | 0.08 |
ENST00000469968.1
ENST00000436927.1 ENST00000469075.1 ENST00000488718.1 ENST00000482491.1 ENST00000340871.4 ENST00000492281.1 ENST00000361463.3 ENST00000391875.2 ENST00000461546.1 |
NVL
|
nuclear VCP-like |
| chr14_-_67981870 | 0.08 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr7_+_99775520 | 0.08 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr11_-_94964210 | 0.08 |
ENST00000416495.2
ENST00000393234.1 |
SESN3
|
sestrin 3 |
| chr19_+_39971505 | 0.08 |
ENST00000544017.1
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr1_+_100435535 | 0.08 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr19_-_55672037 | 0.08 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr11_-_94964354 | 0.07 |
ENST00000536441.1
|
SESN3
|
sestrin 3 |
| chr12_-_6982442 | 0.07 |
ENST00000523102.1
ENST00000524270.1 ENST00000519357.1 |
SPSB2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr19_-_10764509 | 0.07 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr7_+_100450328 | 0.07 |
ENST00000540482.1
ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9
|
solute carrier family 12, member 9 |
| chr7_+_96747030 | 0.07 |
ENST00000360382.4
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chr11_-_102962929 | 0.07 |
ENST00000260247.5
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr6_-_11779403 | 0.07 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr9_-_131038214 | 0.07 |
ENST00000609374.1
|
GOLGA2
|
golgin A2 |
| chr14_-_74551172 | 0.07 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr4_-_10117949 | 0.07 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr2_-_128785688 | 0.07 |
ENST00000259234.6
|
SAP130
|
Sin3A-associated protein, 130kDa |
| chr15_+_75575176 | 0.07 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chr10_+_104678102 | 0.07 |
ENST00000433628.2
|
CNNM2
|
cyclin M2 |
| chr21_+_35445811 | 0.07 |
ENST00000399312.2
|
MRPS6
|
mitochondrial ribosomal protein S6 |
| chr16_-_28936007 | 0.07 |
ENST00000568703.1
ENST00000567483.1 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr1_-_45805607 | 0.07 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr19_+_36236514 | 0.07 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr10_+_120789223 | 0.06 |
ENST00000425699.1
|
NANOS1
|
nanos homolog 1 (Drosophila) |
| chr1_+_100436065 | 0.06 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr5_+_111496554 | 0.06 |
ENST00000442823.2
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chr14_+_24867992 | 0.06 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr12_+_124457670 | 0.06 |
ENST00000539644.1
|
ZNF664
|
zinc finger protein 664 |
| chr6_-_11779014 | 0.06 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr12_+_56521840 | 0.06 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr9_-_116163400 | 0.06 |
ENST00000277315.5
ENST00000448137.1 ENST00000409155.3 |
ALAD
|
aminolevulinate dehydratase |
| chr14_+_54976603 | 0.06 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr12_+_121124599 | 0.06 |
ENST00000228506.3
|
MLEC
|
malectin |
| chr16_-_89883015 | 0.06 |
ENST00000563673.1
ENST00000389301.3 ENST00000568369.1 ENST00000534992.1 ENST00000389302.3 ENST00000543736.1 |
FANCA
|
Fanconi anemia, complementation group A |
| chr9_-_179018 | 0.06 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr19_-_36236292 | 0.06 |
ENST00000378975.3
ENST00000412391.2 ENST00000292879.5 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr9_+_35605274 | 0.06 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr19_-_44100275 | 0.06 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr11_+_129939779 | 0.06 |
ENST00000533195.1
ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr12_+_6982725 | 0.05 |
ENST00000433346.1
|
LRRC23
|
leucine rich repeat containing 23 |
| chr17_-_36956155 | 0.05 |
ENST00000269554.3
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr19_+_36632204 | 0.05 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr17_+_17380294 | 0.05 |
ENST00000268711.3
ENST00000580462.1 |
MED9
|
mediator complex subunit 9 |
| chr9_-_69262509 | 0.05 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr17_+_38296576 | 0.05 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr3_-_14220068 | 0.05 |
ENST00000449060.2
ENST00000511155.1 |
XPC
|
xeroderma pigmentosum, complementation group C |
| chr2_+_105654441 | 0.05 |
ENST00000258455.3
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr2_+_239335449 | 0.05 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chrX_+_54835493 | 0.05 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr10_+_104005272 | 0.05 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr3_+_158519654 | 0.05 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr16_+_3074002 | 0.05 |
ENST00000326266.8
ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6
|
THO complex 6 homolog (Drosophila) |
| chr2_+_71295717 | 0.05 |
ENST00000418807.3
ENST00000443872.2 |
NAGK
|
N-acetylglucosamine kinase |
| chr7_-_139168402 | 0.05 |
ENST00000393039.2
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr12_+_58120044 | 0.05 |
ENST00000542466.2
|
AGAP2-AS1
|
AGAP2 antisense RNA 1 |
| chr1_-_220101944 | 0.05 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr19_-_19729725 | 0.05 |
ENST00000251203.9
|
PBX4
|
pre-B-cell leukemia homeobox 4 |
| chr1_-_223316577 | 0.05 |
ENST00000407096.2
|
TLR5
|
toll-like receptor 5 |
| chr5_-_32313082 | 0.05 |
ENST00000382142.3
|
MTMR12
|
myotubularin related protein 12 |
| chr21_+_44394742 | 0.05 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr6_-_105307417 | 0.05 |
ENST00000524020.1
|
HACE1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr19_+_50887585 | 0.04 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr20_+_18568537 | 0.04 |
ENST00000377452.3
|
DTD1
|
D-tyrosyl-tRNA deacylase 1 |
| chr19_+_49403562 | 0.04 |
ENST00000407032.1
ENST00000452087.1 ENST00000411700.1 |
NUCB1
|
nucleobindin 1 |
| chr6_-_11779174 | 0.04 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr5_+_154238042 | 0.04 |
ENST00000519211.1
ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr3_+_184529929 | 0.04 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr3_+_184529948 | 0.04 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr2_-_160143059 | 0.04 |
ENST00000392796.3
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr15_-_74988281 | 0.04 |
ENST00000566828.1
ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr20_+_60813535 | 0.04 |
ENST00000358053.2
ENST00000313733.3 ENST00000439951.2 |
OSBPL2
|
oxysterol binding protein-like 2 |
| chr9_-_131038266 | 0.04 |
ENST00000490628.1
ENST00000421699.2 ENST00000450617.1 |
GOLGA2
|
golgin A2 |
| chr17_+_19281034 | 0.04 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr2_+_71295766 | 0.04 |
ENST00000533981.1
|
NAGK
|
N-acetylglucosamine kinase |
| chr17_+_42264395 | 0.04 |
ENST00000587989.1
ENST00000590235.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr2_-_160143084 | 0.04 |
ENST00000409990.3
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr17_+_73008755 | 0.04 |
ENST00000584208.1
ENST00000301585.5 |
ICT1
|
immature colon carcinoma transcript 1 |
| chr1_+_15573757 | 0.03 |
ENST00000358897.4
ENST00000417793.1 ENST00000375999.3 ENST00000433640.2 |
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr4_+_9174899 | 0.03 |
ENST00000432515.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr7_-_91764108 | 0.03 |
ENST00000450723.1
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr19_+_10654561 | 0.03 |
ENST00000309469.4
|
ATG4D
|
autophagy related 4D, cysteine peptidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.4 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.5 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.2 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.2 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.1 | 0.2 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 1.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.2 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.5 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.4 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.2 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.1 | 0.2 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.1 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.0 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0052811 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |