Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
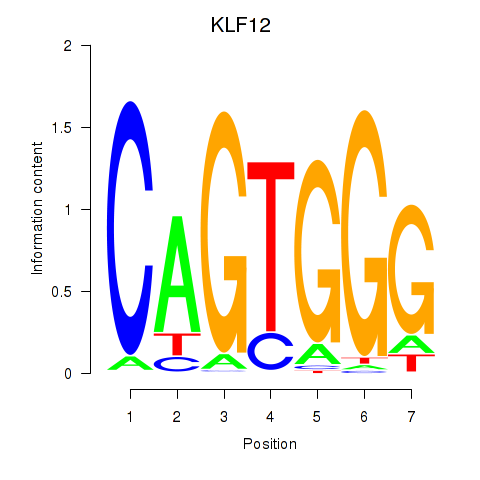
Results for KLF12
Z-value: 1.35
Transcription factors associated with KLF12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF12
|
ENSG00000118922.12 | Kruppel like factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF12 | hg19_v2_chr13_-_74708372_74708409 | 0.86 | 1.4e-01 | Click! |
Activity profile of KLF12 motif
Sorted Z-values of KLF12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153750622 | 1.00 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr12_-_56120838 | 0.66 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr22_-_39268308 | 0.58 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr4_-_5710257 | 0.57 |
ENST00000344938.1
ENST00000344408.5 |
EVC2
|
Ellis van Creveld syndrome 2 |
| chr17_+_27071002 | 0.51 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr12_-_56120865 | 0.50 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr6_-_107436473 | 0.48 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr17_+_1665253 | 0.46 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr5_-_139930713 | 0.44 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr4_+_3443614 | 0.42 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr17_+_4618734 | 0.42 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr12_-_56881874 | 0.41 |
ENST00000539272.1
|
GLS2
|
glutaminase 2 (liver, mitochondrial) |
| chr19_-_5785630 | 0.40 |
ENST00000586012.1
ENST00000590343.1 |
CTB-54O9.9
DUS3L
|
Uncharacterized protein dihydrouridine synthase 3-like (S. cerevisiae) |
| chr17_-_41174367 | 0.39 |
ENST00000587173.1
|
VAT1
|
vesicle amine transport 1 |
| chr2_+_231902193 | 0.39 |
ENST00000373640.4
|
C2orf72
|
chromosome 2 open reading frame 72 |
| chr16_-_67978016 | 0.37 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr14_-_104181771 | 0.37 |
ENST00000554913.1
ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr17_-_36891830 | 0.36 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr22_-_39268192 | 0.36 |
ENST00000216083.6
|
CBX6
|
chromobox homolog 6 |
| chr14_+_65171315 | 0.35 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_-_74779744 | 0.35 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr15_+_40763150 | 0.35 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr18_+_56532053 | 0.35 |
ENST00000592452.1
|
ZNF532
|
zinc finger protein 532 |
| chr7_+_65552756 | 0.35 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr7_+_66800928 | 0.34 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr10_-_77161004 | 0.34 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chr11_+_117070037 | 0.34 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr14_+_63671105 | 0.33 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr2_-_85828867 | 0.32 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr12_+_7060414 | 0.32 |
ENST00000538715.1
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr14_+_75894391 | 0.31 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr20_+_44637526 | 0.30 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr17_-_28257080 | 0.30 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr8_-_30670053 | 0.30 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr3_-_61237050 | 0.30 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr18_+_29598335 | 0.29 |
ENST00000217740.3
|
RNF125
|
ring finger protein 125, E3 ubiquitin protein ligase |
| chr8_-_71519889 | 0.29 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr21_+_38338737 | 0.29 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr16_+_28875126 | 0.29 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr17_-_72864739 | 0.29 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr9_+_131445703 | 0.28 |
ENST00000454747.1
|
SET
|
SET nuclear oncogene |
| chr14_+_70234642 | 0.28 |
ENST00000555349.1
ENST00000554021.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr15_+_90118685 | 0.28 |
ENST00000268138.7
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr5_-_81046841 | 0.28 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_+_33168597 | 0.27 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr2_+_128180842 | 0.27 |
ENST00000402125.2
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr17_-_47045949 | 0.27 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr8_-_42698292 | 0.26 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr11_-_57089671 | 0.26 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr8_-_25281747 | 0.26 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr10_+_98741041 | 0.26 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr19_+_10812108 | 0.25 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr17_+_28256874 | 0.25 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr9_-_35754253 | 0.25 |
ENST00000436428.2
|
MSMP
|
microseminoprotein, prostate associated |
| chr15_+_90118723 | 0.25 |
ENST00000560985.1
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr11_-_22647350 | 0.25 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr17_+_1665345 | 0.25 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_3978083 | 0.25 |
ENST00000600794.1
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr22_-_30198075 | 0.24 |
ENST00000411532.1
|
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr5_+_34656450 | 0.24 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr1_-_55341551 | 0.23 |
ENST00000537443.1
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr18_+_52385068 | 0.23 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr19_+_45174724 | 0.23 |
ENST00000358777.4
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr14_+_65171099 | 0.23 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr12_+_120740119 | 0.23 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr7_-_43965937 | 0.23 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr8_-_144512576 | 0.23 |
ENST00000333480.2
|
MAFA
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
| chr11_-_64856497 | 0.23 |
ENST00000524632.1
ENST00000530719.1 |
AP003068.6
|
transmembrane protein 262 |
| chr17_-_27277615 | 0.22 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr6_+_33168637 | 0.22 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr14_-_61190754 | 0.22 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr12_-_120907374 | 0.22 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr17_-_73840415 | 0.22 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr14_+_52313833 | 0.22 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr19_-_41220957 | 0.22 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr16_+_28875268 | 0.22 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr11_-_62474803 | 0.21 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr19_+_46000479 | 0.21 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr3_+_50316458 | 0.21 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr11_-_64013288 | 0.21 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr10_+_104404644 | 0.21 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr2_-_74710078 | 0.21 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr16_+_29819372 | 0.21 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_-_27593180 | 0.21 |
ENST00000493344.2
ENST00000445933.2 |
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr22_-_42310570 | 0.20 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr10_-_52645416 | 0.20 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr3_-_167813132 | 0.20 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr12_+_121416489 | 0.20 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr14_+_104182105 | 0.20 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr16_-_30798492 | 0.20 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr12_-_56122220 | 0.20 |
ENST00000552692.1
|
CD63
|
CD63 molecule |
| chr19_+_5914213 | 0.20 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr16_+_67360712 | 0.20 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr14_+_32546274 | 0.20 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr22_+_47016277 | 0.20 |
ENST00000406902.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr1_-_19229014 | 0.20 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr11_+_71164149 | 0.19 |
ENST00000319023.2
|
NADSYN1
|
NAD synthetase 1 |
| chr3_-_8686479 | 0.19 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr12_-_56122124 | 0.19 |
ENST00000552754.1
|
CD63
|
CD63 molecule |
| chr12_+_53848549 | 0.19 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr15_-_78112553 | 0.19 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr17_+_7211280 | 0.19 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr1_+_206730484 | 0.19 |
ENST00000304534.8
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr1_+_28918712 | 0.18 |
ENST00000373826.3
|
RAB42
|
RAB42, member RAS oncogene family |
| chr3_-_49722523 | 0.18 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr6_-_33160231 | 0.18 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr3_+_29322803 | 0.18 |
ENST00000396583.3
ENST00000383767.2 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr14_-_55878538 | 0.18 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr17_-_26989136 | 0.18 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr17_-_27418537 | 0.18 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr11_-_568369 | 0.18 |
ENST00000534540.1
ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG
|
MIR210 host gene (non-protein coding) |
| chrX_-_129299847 | 0.17 |
ENST00000319908.3
ENST00000287295.3 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr12_+_53848505 | 0.17 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr12_-_56121580 | 0.17 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr12_+_50366620 | 0.17 |
ENST00000315520.5
|
AQP6
|
aquaporin 6, kidney specific |
| chr19_+_45174994 | 0.17 |
ENST00000403660.3
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr22_+_25202232 | 0.17 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr16_-_103572 | 0.17 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr22_-_37640456 | 0.17 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr22_-_31742218 | 0.16 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr12_+_7060508 | 0.16 |
ENST00000541698.1
ENST00000542462.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr14_+_77425972 | 0.16 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr8_-_107782463 | 0.16 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr13_-_30160925 | 0.16 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_+_121416340 | 0.16 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr1_+_17866290 | 0.16 |
ENST00000361221.3
ENST00000452522.1 ENST00000434513.1 |
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr9_+_131084815 | 0.16 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr16_-_790887 | 0.16 |
ENST00000540986.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr20_-_52687059 | 0.16 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr16_+_103816 | 0.16 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr1_-_114447683 | 0.16 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr1_-_155211017 | 0.16 |
ENST00000536770.1
ENST00000368373.3 |
GBA
|
glucosidase, beta, acid |
| chr12_+_54379569 | 0.16 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr2_-_65659762 | 0.16 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr14_+_104182061 | 0.16 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr1_+_116915270 | 0.16 |
ENST00000418797.1
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr8_+_7705398 | 0.16 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr19_+_49109990 | 0.16 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr1_+_55181490 | 0.15 |
ENST00000371281.3
|
TTC4
|
tetratricopeptide repeat domain 4 |
| chr11_-_2160180 | 0.15 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr15_-_90039805 | 0.15 |
ENST00000544600.1
ENST00000268122.4 |
RHCG
|
Rh family, C glycoprotein |
| chrX_-_129299638 | 0.15 |
ENST00000535724.1
ENST00000346424.2 |
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr20_+_8112824 | 0.15 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr16_-_790982 | 0.15 |
ENST00000301694.5
ENST00000251588.2 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr8_-_7320974 | 0.15 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr17_-_79827808 | 0.15 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chrX_-_54209640 | 0.15 |
ENST00000375180.2
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr11_-_61129306 | 0.15 |
ENST00000544118.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr6_-_31864977 | 0.15 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr17_-_4889508 | 0.15 |
ENST00000574606.2
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr2_+_105471969 | 0.15 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr10_+_81272287 | 0.15 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr8_+_38644715 | 0.15 |
ENST00000317827.4
ENST00000379931.3 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr2_+_27593389 | 0.15 |
ENST00000233575.2
ENST00000543024.1 ENST00000537606.1 |
SNX17
|
sorting nexin 17 |
| chr19_+_6772710 | 0.15 |
ENST00000304076.2
ENST00000602142.1 ENST00000596764.1 |
VAV1
|
vav 1 guanine nucleotide exchange factor |
| chr1_-_153600656 | 0.15 |
ENST00000339556.4
ENST00000440685.2 |
S100A13
|
S100 calcium binding protein A13 |
| chr6_-_30640761 | 0.15 |
ENST00000415603.1
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr2_-_27593306 | 0.15 |
ENST00000347454.4
|
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr12_-_24737089 | 0.14 |
ENST00000483544.1
|
LINC00477
|
long intergenic non-protein coding RNA 477 |
| chr6_+_43612750 | 0.14 |
ENST00000372165.4
ENST00000372163.4 |
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas) |
| chr8_+_145086574 | 0.14 |
ENST00000377470.3
ENST00000447830.2 |
SPATC1
|
spermatogenesis and centriole associated 1 |
| chr22_-_24110063 | 0.14 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr15_+_31619013 | 0.14 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr1_+_27561104 | 0.14 |
ENST00000361771.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr11_-_125365435 | 0.14 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr1_+_156084461 | 0.14 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr12_+_118573663 | 0.14 |
ENST00000261313.2
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr16_+_3019246 | 0.14 |
ENST00000318782.8
ENST00000293978.8 |
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr7_+_140373619 | 0.14 |
ENST00000483369.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr8_+_38644778 | 0.14 |
ENST00000276520.8
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr6_+_35995531 | 0.14 |
ENST00000229794.4
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr11_-_118966167 | 0.13 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr10_-_52645379 | 0.13 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr11_+_61159832 | 0.13 |
ENST00000334888.5
ENST00000398979.3 |
TMEM216
|
transmembrane protein 216 |
| chr17_+_20059358 | 0.13 |
ENST00000536879.1
ENST00000395522.2 ENST00000395525.3 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr16_+_30671223 | 0.13 |
ENST00000568722.1
|
FBRS
|
fibrosin |
| chr17_-_73840774 | 0.13 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr6_-_134638767 | 0.13 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_-_64673665 | 0.13 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr10_-_71906342 | 0.13 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr1_-_51796226 | 0.13 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr17_+_7155343 | 0.13 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr17_+_1633755 | 0.13 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr17_-_39156138 | 0.13 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr12_+_7060432 | 0.13 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr14_+_23564700 | 0.12 |
ENST00000554203.1
|
C14orf119
|
chromosome 14 open reading frame 119 |
| chrX_-_45060135 | 0.12 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr11_-_1643368 | 0.12 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr19_-_56109119 | 0.12 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr7_-_65447192 | 0.12 |
ENST00000421103.1
ENST00000345660.6 ENST00000304895.4 |
GUSB
|
glucuronidase, beta |
| chr3_-_182703688 | 0.12 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr5_-_180632147 | 0.12 |
ENST00000274773.7
|
TRIM7
|
tripartite motif containing 7 |
| chr1_+_26798955 | 0.12 |
ENST00000361427.5
|
HMGN2
|
high mobility group nucleosomal binding domain 2 |
| chr3_-_133614467 | 0.12 |
ENST00000469959.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr4_+_103422499 | 0.12 |
ENST00000511926.1
ENST00000507079.1 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr4_+_95972822 | 0.12 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_-_32687923 | 0.12 |
ENST00000309777.6
ENST00000344461.3 ENST00000373593.1 ENST00000545122.1 |
TMEM234
|
transmembrane protein 234 |
| chr1_+_86064794 | 0.12 |
ENST00000426794.1
|
RP11-290M5.2
|
RP11-290M5.2 |
| chr3_+_184037466 | 0.12 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_153600869 | 0.12 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF12
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.1 | 1.7 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.7 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.4 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.4 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.1 | 0.3 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of fertilization(GO:0080154) activation of meiosis(GO:0090427) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.3 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.4 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.0 | 0.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0031947 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.0 | 0.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:2000767 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0072300 | positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.0 | 0.0 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0061086 | regulation of mRNA export from nucleus(GO:0010793) negative regulation of histone H3-K27 methylation(GO:0061086) chromatin maintenance(GO:0070827) |
| 0.0 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.3 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.2 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |