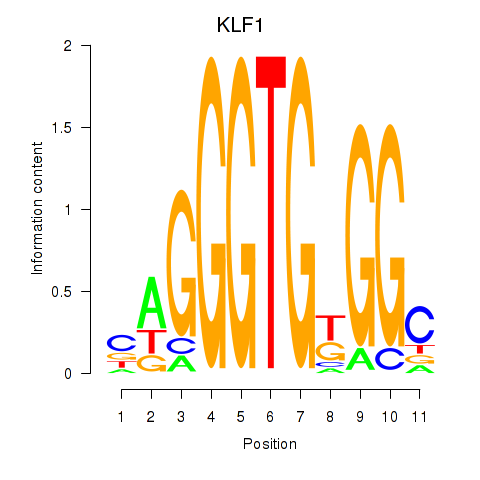
|
chr14_+_32414059
Show fit
|
1.23 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein
|
|
chr20_-_52687059
Show fit
|
0.75 |
ENST00000371435.2
ENST00000395961.3
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr16_-_67427389
Show fit
|
0.62 |
ENST00000562206.1
ENST00000290942.5
ENST00000393957.2
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr3_+_186330712
Show fit
|
0.56 |
ENST00000411641.2
ENST00000273784.5
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr5_+_162864575
Show fit
|
0.53 |
ENST00000512163.1
ENST00000393929.1
ENST00000340828.2
ENST00000511683.2
ENST00000510097.1
ENST00000511490.2
ENST00000510664.1
|
CCNG1
|
cyclin G1
|
|
chr1_-_33168336
Show fit
|
0.50 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr16_-_31161380
Show fit
|
0.49 |
ENST00000569305.1
ENST00000418068.2
ENST00000268281.4
|
PRSS36
|
protease, serine, 36
|
|
chr22_-_20138302
Show fit
|
0.48 |
ENST00000540078.1
ENST00000439765.2
|
AC006547.14
|
uncharacterized protein LOC388849
|
|
chr8_-_101963482
Show fit
|
0.45 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta
|
|
chr1_+_6845578
Show fit
|
0.45 |
ENST00000467404.2
ENST00000439411.2
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr7_-_91875109
Show fit
|
0.43 |
ENST00000412043.2
ENST00000430102.1
ENST00000425073.1
ENST00000394503.2
ENST00000454017.1
ENST00000440209.1
ENST00000413688.1
ENST00000452773.1
ENST00000433016.1
ENST00000394505.2
ENST00000422347.1
ENST00000458493.1
ENST00000425919.1
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr8_+_120428546
Show fit
|
0.42 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed
|
|
chrX_+_47077680
Show fit
|
0.42 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr2_-_38604398
Show fit
|
0.40 |
ENST00000443098.1
ENST00000449130.1
ENST00000378954.4
ENST00000539122.1
ENST00000419554.2
ENST00000451483.1
ENST00000406122.1
|
ATL2
|
atlastin GTPase 2
|
|
chr6_+_43968306
Show fit
|
0.40 |
ENST00000442114.2
ENST00000336600.5
ENST00000439969.2
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr5_+_95998673
Show fit
|
0.38 |
ENST00000514845.1
|
CAST
|
calpastatin
|
|
chr9_-_33264676
Show fit
|
0.38 |
ENST00000472232.3
ENST00000379704.2
|
BAG1
|
BCL2-associated athanogene
|
|
chr9_-_33264557
Show fit
|
0.37 |
ENST00000473781.1
ENST00000488499.1
|
BAG1
|
BCL2-associated athanogene
|
|
chr2_+_206950095
Show fit
|
0.37 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3
|
|
chr18_-_48723690
Show fit
|
0.36 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C
|
|
chr11_+_62475130
Show fit
|
0.35 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr20_-_62710832
Show fit
|
0.35 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr14_+_59951161
Show fit
|
0.34 |
ENST00000261247.9
ENST00000425728.2
ENST00000556985.1
ENST00000554271.1
ENST00000554795.1
|
JKAMP
|
JNK1/MAPK8-associated membrane protein
|
|
chr17_-_7080227
Show fit
|
0.33 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr8_-_143867946
Show fit
|
0.33 |
ENST00000301263.4
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr8_+_38614754
Show fit
|
0.31 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr1_-_244615425
Show fit
|
0.31 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase
|
|
chr12_-_57443886
Show fit
|
0.31 |
ENST00000300119.3
|
MYO1A
|
myosin IA
|
|
chr19_+_38880695
Show fit
|
0.30 |
ENST00000587947.1
ENST00000338502.4
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr9_+_74764340
Show fit
|
0.30 |
ENST00000376986.1
ENST00000358399.3
|
GDA
|
guanine deaminase
|
|
chr1_-_185286461
Show fit
|
0.30 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein
|
|
chr5_+_95998714
Show fit
|
0.29 |
ENST00000506811.1
ENST00000514055.1
|
CAST
|
calpastatin
|
|
chr6_-_138428613
Show fit
|
0.29 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr17_+_40950797
Show fit
|
0.29 |
ENST00000588408.1
ENST00000585355.1
|
CNTD1
|
cyclin N-terminal domain containing 1
|
|
chr2_-_55237484
Show fit
|
0.29 |
ENST00000394609.2
|
RTN4
|
reticulon 4
|
|
chr17_+_46131912
Show fit
|
0.28 |
ENST00000584634.1
ENST00000580050.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1
|
|
chr12_+_56477093
Show fit
|
0.28 |
ENST00000549672.1
ENST00000415288.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3
|
|
chr19_+_7599792
Show fit
|
0.27 |
ENST00000600942.1
ENST00000593924.1
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr7_+_35840819
Show fit
|
0.27 |
ENST00000399035.3
|
SEPT7
|
septin 7
|
|
chr2_+_67624430
Show fit
|
0.27 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1
|
|
chr19_-_40440533
Show fit
|
0.26 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein
|
|
chr6_-_82462425
Show fit
|
0.26 |
ENST00000369754.3
ENST00000320172.6
ENST00000369756.3
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr3_-_113464906
Show fit
|
0.26 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr17_+_46131843
Show fit
|
0.26 |
ENST00000577411.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1
|
|
chr15_+_76352178
Show fit
|
0.25 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr17_-_27405875
Show fit
|
0.25 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr5_+_110074685
Show fit
|
0.25 |
ENST00000355943.3
ENST00000447245.2
|
SLC25A46
|
solute carrier family 25, member 46
|
|
chr5_-_89770582
Show fit
|
0.25 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2
|
|
chr19_+_1077393
Show fit
|
0.25 |
ENST00000590577.1
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr15_+_90931450
Show fit
|
0.25 |
ENST00000268182.5
ENST00000560738.1
ENST00000560418.1
|
IQGAP1
|
IQ motif containing GTPase activating protein 1
|
|
chr8_-_21988558
Show fit
|
0.25 |
ENST00000312841.8
|
HR
|
hair growth associated
|
|
chr5_+_82767583
Show fit
|
0.25 |
ENST00000512590.2
ENST00000513960.1
ENST00000513984.1
ENST00000502527.2
|
VCAN
|
versican
|
|
chr7_-_91875358
Show fit
|
0.24 |
ENST00000458177.1
ENST00000394507.1
ENST00000340022.2
ENST00000444960.1
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr19_-_51523275
Show fit
|
0.24 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr4_-_103748271
Show fit
|
0.24 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr1_-_59165763
Show fit
|
0.24 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1
|
|
chr19_+_18451439
Show fit
|
0.24 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I
|
|
chr14_+_21569245
Show fit
|
0.24 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253
|
|
chr5_+_95998070
Show fit
|
0.24 |
ENST00000421689.2
ENST00000510756.1
ENST00000512620.1
|
CAST
|
calpastatin
|
|
chr1_+_62901968
Show fit
|
0.23 |
ENST00000452143.1
ENST00000442679.1
ENST00000371146.1
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr6_-_84140757
Show fit
|
0.23 |
ENST00000541327.1
ENST00000369705.3
ENST00000543031.1
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr3_-_101395936
Show fit
|
0.23 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr20_-_52687030
Show fit
|
0.23 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr10_+_17272608
Show fit
|
0.23 |
ENST00000421459.2
|
VIM
|
vimentin
|
|
chr5_+_95998746
Show fit
|
0.23 |
ENST00000508608.1
|
CAST
|
calpastatin
|
|
chr6_+_42952237
Show fit
|
0.23 |
ENST00000485511.1
ENST00000394110.3
ENST00000472118.1
ENST00000461010.1
|
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta
|
|
chr2_+_173292301
Show fit
|
0.23 |
ENST00000264106.6
ENST00000375221.2
ENST00000343713.4
|
ITGA6
|
integrin, alpha 6
|
|
chr3_-_50383096
Show fit
|
0.22 |
ENST00000442887.1
ENST00000360165.3
|
ZMYND10
|
zinc finger, MYND-type containing 10
|
|
chr5_+_133562095
Show fit
|
0.22 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3
|
|
chr1_+_222885884
Show fit
|
0.22 |
ENST00000340934.5
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr10_+_98592009
Show fit
|
0.22 |
ENST00000540664.1
ENST00000371103.3
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr12_+_69004805
Show fit
|
0.22 |
ENST00000541216.1
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr12_+_100661156
Show fit
|
0.22 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae)
|
|
chr7_+_17338239
Show fit
|
0.21 |
ENST00000242057.4
|
AHR
|
aryl hydrocarbon receptor
|
|
chr11_-_71781096
Show fit
|
0.21 |
ENST00000535087.1
ENST00000535838.1
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr6_+_74405804
Show fit
|
0.21 |
ENST00000287097.5
|
CD109
|
CD109 molecule
|
|
chr17_+_34900737
Show fit
|
0.21 |
ENST00000304718.4
ENST00000485685.2
|
GGNBP2
|
gametogenetin binding protein 2
|
|
chr6_+_74405501
Show fit
|
0.21 |
ENST00000437994.2
ENST00000422508.2
|
CD109
|
CD109 molecule
|
|
chr4_+_178230985
Show fit
|
0.21 |
ENST00000264596.3
|
NEIL3
|
nei endonuclease VIII-like 3 (E. coli)
|
|
chr3_+_181429704
Show fit
|
0.20 |
ENST00000431565.2
ENST00000325404.1
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr1_-_98510843
Show fit
|
0.20 |
ENST00000413670.2
ENST00000538428.1
|
MIR137HG
|
MIR137 host gene (non-protein coding)
|
|
chr3_-_88108192
Show fit
|
0.20 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr7_-_102158157
Show fit
|
0.20 |
ENST00000541662.1
ENST00000306682.6
ENST00000465829.1
|
RASA4B
|
RAS p21 protein activator 4B
|
|
chr5_+_102595119
Show fit
|
0.20 |
ENST00000510890.1
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr20_-_62168672
Show fit
|
0.20 |
ENST00000217185.2
|
PTK6
|
protein tyrosine kinase 6
|
|
chr12_+_116997186
Show fit
|
0.20 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2
|
|
chr6_+_142623063
Show fit
|
0.20 |
ENST00000296932.8
ENST00000367609.3
|
GPR126
|
G protein-coupled receptor 126
|
|
chr1_+_6845497
Show fit
|
0.20 |
ENST00000473578.1
ENST00000557126.1
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr1_+_29213584
Show fit
|
0.19 |
ENST00000343067.4
ENST00000356093.2
ENST00000398863.2
ENST00000373800.3
ENST00000349460.4
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr19_-_51523412
Show fit
|
0.19 |
ENST00000391805.1
ENST00000599077.1
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr16_-_30125177
Show fit
|
0.19 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr19_-_36297348
Show fit
|
0.19 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr4_+_129730947
Show fit
|
0.19 |
ENST00000452328.2
ENST00000504089.1
|
PHF17
|
jade family PHD finger 1
|
|
chr10_+_94608245
Show fit
|
0.19 |
ENST00000443748.2
ENST00000260762.6
|
EXOC6
|
exocyst complex component 6
|
|
chr1_+_224803995
Show fit
|
0.19 |
ENST00000272133.3
|
CNIH3
|
cornichon family AMPA receptor auxiliary protein 3
|
|
chr8_+_38758737
Show fit
|
0.19 |
ENST00000521746.1
ENST00000420274.1
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr8_-_101963677
Show fit
|
0.19 |
ENST00000395956.3
ENST00000395953.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta
|
|
chr1_+_97187318
Show fit
|
0.19 |
ENST00000609116.1
ENST00000370198.1
ENST00000370197.1
ENST00000426398.2
ENST00000394184.3
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr1_+_62902308
Show fit
|
0.19 |
ENST00000339950.4
|
USP1
|
ubiquitin specific peptidase 1
|
|
chr1_-_205744205
Show fit
|
0.18 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr14_-_59950724
Show fit
|
0.18 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-)
|
|
chr2_+_28974603
Show fit
|
0.18 |
ENST00000441461.1
ENST00000358506.2
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr8_+_38758845
Show fit
|
0.18 |
ENST00000519640.1
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr13_+_88324870
Show fit
|
0.18 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr2_-_148779106
Show fit
|
0.18 |
ENST00000416719.1
ENST00000264169.2
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr6_-_30710265
Show fit
|
0.18 |
ENST00000438162.1
ENST00000454845.1
|
FLOT1
|
flotillin 1
|
|
chr1_-_115259337
Show fit
|
0.18 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr3_+_106959530
Show fit
|
0.18 |
ENST00000466734.1
ENST00000463143.1
ENST00000490441.1
|
LINC00883
|
long intergenic non-protein coding RNA 883
|
|
chr1_+_111682827
Show fit
|
0.18 |
ENST00000357172.4
|
CEPT1
|
choline/ethanolamine phosphotransferase 1
|
|
chr14_-_65409438
Show fit
|
0.18 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr1_-_166135952
Show fit
|
0.18 |
ENST00000354422.3
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr19_-_36523529
Show fit
|
0.18 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3
|
|
chr20_+_43029911
Show fit
|
0.18 |
ENST00000443598.2
ENST00000316099.4
ENST00000415691.2
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr5_-_37249397
Show fit
|
0.18 |
ENST00000425232.2
ENST00000274258.7
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr1_-_39395165
Show fit
|
0.18 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr15_-_64648273
Show fit
|
0.17 |
ENST00000607537.1
ENST00000303052.7
ENST00000303032.6
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr7_-_102257139
Show fit
|
0.17 |
ENST00000521076.1
ENST00000462172.1
ENST00000522801.1
ENST00000449970.2
ENST00000262940.7
|
RASA4
|
RAS p21 protein activator 4
|
|
chr6_+_142622991
Show fit
|
0.17 |
ENST00000230173.6
ENST00000367608.2
|
GPR126
|
G protein-coupled receptor 126
|
|
chr16_+_30675654
Show fit
|
0.17 |
ENST00000287468.5
ENST00000395073.2
|
FBRS
|
fibrosin
|
|
chr14_-_65409502
Show fit
|
0.17 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr3_-_186080012
Show fit
|
0.17 |
ENST00000544847.1
ENST00000265022.3
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr3_-_182817297
Show fit
|
0.17 |
ENST00000539926.1
ENST00000476176.1
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha)
|
|
chr1_+_154966058
Show fit
|
0.17 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein
|
|
chr20_+_47662805
Show fit
|
0.17 |
ENST00000262982.2
ENST00000542325.1
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr9_+_139746792
Show fit
|
0.17 |
ENST00000317446.2
ENST00000445819.1
|
MAMDC4
|
MAM domain containing 4
|
|
chr12_-_110939870
Show fit
|
0.17 |
ENST00000447578.2
ENST00000546588.1
ENST00000360579.7
ENST00000549970.1
ENST00000549578.1
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae)
|
|
chr3_+_106959552
Show fit
|
0.16 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883
|
|
chr1_+_173837488
Show fit
|
0.16 |
ENST00000427304.1
ENST00000432989.1
ENST00000367702.1
|
ZBTB37
|
zinc finger and BTB domain containing 37
|
|
chr1_+_17914907
Show fit
|
0.16 |
ENST00000375420.3
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr1_+_203765437
Show fit
|
0.16 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6
|
|
chr17_-_7166500
Show fit
|
0.16 |
ENST00000575313.1
ENST00000397317.4
|
CLDN7
|
claudin 7
|
|
chr11_-_1606513
Show fit
|
0.16 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1
|
|
chr16_+_46723552
Show fit
|
0.16 |
ENST00000219097.2
ENST00000568364.2
|
ORC6
|
origin recognition complex, subunit 6
|
|
chr19_-_44008863
Show fit
|
0.16 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr16_+_28996114
Show fit
|
0.16 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells
|
|
chr14_+_96342729
Show fit
|
0.16 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617
|
|
chr11_-_34937858
Show fit
|
0.16 |
ENST00000278359.5
|
APIP
|
APAF1 interacting protein
|
|
chr2_-_55276320
Show fit
|
0.16 |
ENST00000357376.3
|
RTN4
|
reticulon 4
|
|
chr9_+_110046334
Show fit
|
0.16 |
ENST00000416373.2
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr17_-_74023474
Show fit
|
0.16 |
ENST00000301607.3
|
EVPL
|
envoplakin
|
|
chr5_-_137368708
Show fit
|
0.15 |
ENST00000033079.3
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr1_-_205744574
Show fit
|
0.15 |
ENST00000367139.3
ENST00000235932.4
ENST00000437324.2
ENST00000414729.1
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr17_-_27188984
Show fit
|
0.15 |
ENST00000582320.2
|
MIR144
|
microRNA 451b
|
|
chr1_+_67390578
Show fit
|
0.15 |
ENST00000371018.3
ENST00000355977.6
ENST00000357692.2
ENST00000401041.1
ENST00000371016.1
ENST00000371014.1
ENST00000371012.2
|
MIER1
|
mesoderm induction early response 1, transcriptional regulator
|
|
chr13_-_45010939
Show fit
|
0.15 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr2_+_28974531
Show fit
|
0.15 |
ENST00000420282.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr3_+_49449636
Show fit
|
0.15 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered
|
|
chr12_-_80328949
Show fit
|
0.14 |
ENST00000450142.2
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr8_-_103136481
Show fit
|
0.14 |
ENST00000524209.1
ENST00000517822.1
ENST00000523923.1
ENST00000521599.1
ENST00000521964.1
ENST00000311028.3
ENST00000518166.1
|
NCALD
|
neurocalcin delta
|
|
chr2_-_153574480
Show fit
|
0.14 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr2_+_74212073
Show fit
|
0.14 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25
|
|
chr2_-_61765732
Show fit
|
0.14 |
ENST00000443240.1
ENST00000436018.1
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr16_+_30969055
Show fit
|
0.14 |
ENST00000452917.1
|
SETD1A
|
SET domain containing 1A
|
|
chr2_+_173292280
Show fit
|
0.14 |
ENST00000264107.7
|
ITGA6
|
integrin, alpha 6
|
|
chr3_-_182817367
Show fit
|
0.14 |
ENST00000265594.4
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha)
|
|
chr2_-_167232484
Show fit
|
0.14 |
ENST00000375387.4
ENST00000303354.6
ENST00000409672.1
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr15_+_63796779
Show fit
|
0.14 |
ENST00000561442.1
ENST00000560070.1
ENST00000540797.1
ENST00000380324.3
ENST00000268049.7
ENST00000536001.1
ENST00000539772.1
|
USP3
|
ubiquitin specific peptidase 3
|
|
chr16_+_202686
Show fit
|
0.14 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta
|
|
chr12_-_80328700
Show fit
|
0.14 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr11_-_124543725
Show fit
|
0.14 |
ENST00000545756.1
ENST00000263593.3
|
SIAE
|
sialic acid acetylesterase
|
|
chr12_-_112450915
Show fit
|
0.14 |
ENST00000437003.2
ENST00000552374.2
ENST00000550831.3
ENST00000354825.3
ENST00000549537.2
ENST00000355445.3
|
TMEM116
|
transmembrane protein 116
|
|
chr12_+_8185288
Show fit
|
0.14 |
ENST00000162391.3
|
FOXJ2
|
forkhead box J2
|
|
chr11_+_71903169
Show fit
|
0.14 |
ENST00000393676.3
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr6_-_39282221
Show fit
|
0.13 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17
|
|
chr6_-_30709980
Show fit
|
0.13 |
ENST00000416018.1
ENST00000445853.1
ENST00000413165.1
ENST00000418160.1
|
FLOT1
|
flotillin 1
|
|
chr2_-_174828892
Show fit
|
0.13 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor
|
|
chr4_+_129730779
Show fit
|
0.13 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1
|
|
chr6_-_30710447
Show fit
|
0.13 |
ENST00000456573.2
|
FLOT1
|
flotillin 1
|
|
chr15_+_75074410
Show fit
|
0.13 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase
|
|
chr2_+_172378757
Show fit
|
0.13 |
ENST00000409484.1
ENST00000321348.4
ENST00000375252.3
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr4_+_129730839
Show fit
|
0.13 |
ENST00000511647.1
|
PHF17
|
jade family PHD finger 1
|
|
chr5_-_95297534
Show fit
|
0.13 |
ENST00000513343.1
ENST00000431061.2
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr20_+_58515417
Show fit
|
0.13 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B
|
|
chr11_-_6440624
Show fit
|
0.13 |
ENST00000311051.3
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr19_-_15442701
Show fit
|
0.13 |
ENST00000594841.1
ENST00000601941.1
|
BRD4
|
bromodomain containing 4
|
|
chr8_-_101962777
Show fit
|
0.13 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta
|
|
chr7_+_35840542
Show fit
|
0.13 |
ENST00000435235.1
ENST00000399034.2
ENST00000350320.6
ENST00000469679.2
|
SEPT7
|
septin 7
|
|
chr4_+_129731074
Show fit
|
0.13 |
ENST00000512960.1
ENST00000503785.1
ENST00000514740.1
|
PHF17
|
jade family PHD finger 1
|
|
chr6_-_30710510
Show fit
|
0.13 |
ENST00000376389.3
|
FLOT1
|
flotillin 1
|
|
chr15_-_44069513
Show fit
|
0.13 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3
|
|
chr4_-_25865159
Show fit
|
0.13 |
ENST00000502949.1
ENST00000264868.5
ENST00000513691.1
ENST00000514872.1
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr9_+_71736177
Show fit
|
0.13 |
ENST00000606364.1
ENST00000453658.2
|
TJP2
|
tight junction protein 2
|
|
chr12_+_69004736
Show fit
|
0.12 |
ENST00000545720.2
|
RAP1B
|
RAP1B, member of RAS oncogene family
|
|
chr6_+_130339710
Show fit
|
0.12 |
ENST00000526087.1
ENST00000533560.1
ENST00000361794.2
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr2_+_173292390
Show fit
|
0.12 |
ENST00000442250.1
ENST00000458358.1
ENST00000409080.1
|
ITGA6
|
integrin, alpha 6
|
|
chr1_-_243326612
Show fit
|
0.12 |
ENST00000492145.1
ENST00000490813.1
ENST00000464936.1
|
CEP170
|
centrosomal protein 170kDa
|
|
chr4_+_141294628
Show fit
|
0.12 |
ENST00000512749.1
ENST00000608372.1
ENST00000506597.1
ENST00000394201.4
ENST00000510586.1
|
SCOC
|
short coiled-coil protein
|
|
chr19_+_35607166
Show fit
|
0.12 |
ENST00000604255.1
ENST00000346446.5
ENST00000344013.6
ENST00000603449.1
ENST00000406988.1
ENST00000605550.1
ENST00000604804.1
ENST00000605552.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr17_-_7590745
Show fit
|
0.12 |
ENST00000514944.1
ENST00000503591.1
ENST00000455263.2
ENST00000420246.2
ENST00000445888.2
ENST00000509690.1
ENST00000604348.1
ENST00000269305.4
|
TP53
|
tumor protein p53
|
|
chr5_-_130970723
Show fit
|
0.12 |
ENST00000308008.6
ENST00000296859.6
ENST00000507093.1
ENST00000510071.1
ENST00000509018.1
ENST00000307984.5
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr1_+_33231221
Show fit
|
0.12 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522
|
|
chr1_+_156096336
Show fit
|
0.12 |
ENST00000504687.1
ENST00000473598.2
|
LMNA
|
lamin A/C
|
|
chr1_-_109940550
Show fit
|
0.12 |
ENST00000256637.6
|
SORT1
|
sortilin 1
|
|
chr17_-_46716647
Show fit
|
0.12 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17
|
|
chr7_-_16460863
Show fit
|
0.12 |
ENST00000407010.2
ENST00000399310.3
|
ISPD
|
isoprenoid synthase domain containing
|
|
chr17_-_74023291
Show fit
|
0.12 |
ENST00000586740.1
|
EVPL
|
envoplakin
|
|
chr9_-_99329098
Show fit
|
0.12 |
ENST00000452280.1
|
CDC14B
|
cell division cycle 14B
|
|
chr11_-_6440283
Show fit
|
0.12 |
ENST00000299402.6
ENST00000609360.1
ENST00000389906.2
ENST00000532020.2
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr13_-_20437772
Show fit
|
0.11 |
ENST00000337963.4
|
ZMYM5
|
zinc finger, MYM-type 5
|
|
chr19_-_51522955
Show fit
|
0.11 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10
|