Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
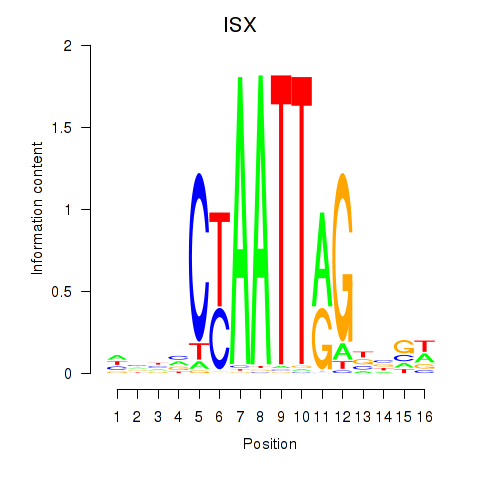
Results for ISX
Z-value: 1.23
Transcription factors associated with ISX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISX
|
ENSG00000175329.8 | intestine specific homeobox |
Activity profile of ISX motif
Sorted Z-values of ISX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_94789663 | 1.77 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr16_+_12059091 | 1.50 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_-_89442940 | 1.39 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr2_-_228244013 | 1.16 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr10_+_91461337 | 1.03 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr1_+_28261492 | 1.03 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_-_197115818 | 0.92 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr17_-_18266818 | 0.89 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr4_+_66536248 | 0.87 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr17_-_57229155 | 0.80 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr19_-_4717835 | 0.77 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr1_-_143913143 | 0.74 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr1_-_112281875 | 0.72 |
ENST00000527621.1
ENST00000534365.1 ENST00000357260.5 |
FAM212B
|
family with sequence similarity 212, member B |
| chr1_+_28261621 | 0.71 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_+_95523621 | 0.68 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr12_+_69201923 | 0.64 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr6_+_31126291 | 0.60 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr11_-_62414070 | 0.60 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr5_-_159846066 | 0.60 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr12_+_16500037 | 0.59 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_3521518 | 0.58 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr6_-_85474219 | 0.58 |
ENST00000369663.5
|
TBX18
|
T-box 18 |
| chr19_-_52307357 | 0.58 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr11_-_95522639 | 0.56 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr1_+_28261533 | 0.55 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr6_-_111927062 | 0.54 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr14_-_88200641 | 0.53 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr4_-_174255536 | 0.53 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr3_-_149095652 | 0.52 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr6_-_111927449 | 0.52 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr10_+_99205959 | 0.52 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr12_+_16500599 | 0.52 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr17_+_59489112 | 0.51 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chrX_+_43515467 | 0.51 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr8_+_38831683 | 0.48 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chrX_+_77166172 | 0.47 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chrY_+_14813160 | 0.47 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr20_+_44441271 | 0.45 |
ENST00000335046.3
ENST00000243893.6 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr6_-_119256279 | 0.45 |
ENST00000316068.3
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr9_+_99212403 | 0.44 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr13_+_40229764 | 0.43 |
ENST00000416691.1
ENST00000422759.2 ENST00000455146.3 |
COG6
|
component of oligomeric golgi complex 6 |
| chr3_+_28390637 | 0.43 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr9_+_104296163 | 0.42 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr12_+_16500571 | 0.41 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr7_-_86849836 | 0.40 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr8_+_110552046 | 0.40 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr1_+_206138457 | 0.39 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr15_-_55700457 | 0.39 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr1_-_159832438 | 0.38 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr20_+_44441215 | 0.38 |
ENST00000356455.4
ENST00000405520.1 |
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr10_+_99205894 | 0.38 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr4_-_141348789 | 0.37 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr7_-_64467031 | 0.37 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr11_+_77532233 | 0.37 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr3_-_15563229 | 0.36 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr14_+_52313833 | 0.36 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr10_-_105110831 | 0.36 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr19_-_47137942 | 0.36 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr5_+_159848807 | 0.36 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr4_-_87515202 | 0.35 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr21_+_35014783 | 0.35 |
ENST00000381291.4
ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr15_+_48413169 | 0.34 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr19_-_51071302 | 0.34 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chrX_+_108779004 | 0.34 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr17_-_40828969 | 0.33 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr14_+_104182105 | 0.32 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr5_-_95297678 | 0.32 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr10_-_1094819 | 0.32 |
ENST00000429642.1
|
IDI1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr14_-_53258314 | 0.32 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr17_-_27418537 | 0.31 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr17_-_40829026 | 0.30 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr6_+_130339710 | 0.30 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_+_120839005 | 0.30 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chrX_+_37639302 | 0.30 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_+_115862858 | 0.29 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr20_+_24449821 | 0.29 |
ENST00000376862.3
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr3_-_129147432 | 0.29 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr11_+_100862811 | 0.29 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr4_-_141348763 | 0.27 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr9_+_131452239 | 0.27 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr5_-_95297534 | 0.26 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chrX_+_77154935 | 0.26 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr12_-_10978957 | 0.25 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr4_-_141348999 | 0.25 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr7_-_86849883 | 0.25 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr16_+_31724552 | 0.24 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr17_-_45266542 | 0.24 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr5_-_98262240 | 0.24 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr15_-_55700522 | 0.23 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr11_-_95522907 | 0.23 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr4_-_69536346 | 0.22 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr8_-_12612962 | 0.22 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr13_+_38923959 | 0.21 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr4_-_171012844 | 0.21 |
ENST00000502392.1
|
AADAT
|
aminoadipate aminotransferase |
| chrX_-_47509994 | 0.20 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chr19_+_42041860 | 0.20 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr20_+_44441304 | 0.20 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr6_+_30687978 | 0.20 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr6_-_32095968 | 0.20 |
ENST00000375203.3
ENST00000375201.4 |
ATF6B
|
activating transcription factor 6 beta |
| chr19_+_36346734 | 0.20 |
ENST00000586102.3
|
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr19_+_13001840 | 0.20 |
ENST00000222214.5
ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH
|
glutaryl-CoA dehydrogenase |
| chr14_-_73925225 | 0.20 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr14_+_39944025 | 0.19 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr17_-_18266660 | 0.19 |
ENST00000582653.1
ENST00000352886.6 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr17_-_18266797 | 0.19 |
ENST00000316694.3
ENST00000539052.1 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr22_-_29137771 | 0.19 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr1_-_150669500 | 0.19 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_+_70574088 | 0.19 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr17_-_18266765 | 0.19 |
ENST00000354098.3
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr4_-_100242549 | 0.18 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr3_+_133292574 | 0.18 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr6_+_64345698 | 0.18 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr13_+_73632897 | 0.18 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr5_+_159848854 | 0.18 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr14_-_51027838 | 0.17 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_39693111 | 0.17 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr6_+_36562132 | 0.17 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr3_+_51851612 | 0.17 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr18_-_67624160 | 0.17 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr11_+_119205222 | 0.17 |
ENST00000311413.4
|
RNF26
|
ring finger protein 26 |
| chr2_+_172778952 | 0.17 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr1_+_62902308 | 0.16 |
ENST00000339950.4
|
USP1
|
ubiquitin specific peptidase 1 |
| chr10_-_104001231 | 0.16 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr3_-_48659193 | 0.16 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr20_-_50418972 | 0.16 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr9_+_136501478 | 0.15 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr12_-_120966943 | 0.15 |
ENST00000552443.1
ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5
|
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr6_-_119256311 | 0.15 |
ENST00000316316.6
|
MCM9
|
minichromosome maintenance complex component 9 |
| chr1_-_150849208 | 0.15 |
ENST00000358595.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr5_+_140557371 | 0.15 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr19_+_42041702 | 0.15 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr3_+_37284668 | 0.15 |
ENST00000361924.2
ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4
|
golgin A4 |
| chr17_-_38821373 | 0.15 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_+_162336686 | 0.14 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr6_-_135271219 | 0.14 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr15_-_75199213 | 0.14 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr4_-_70080449 | 0.13 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr8_+_110551925 | 0.13 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr7_+_142985308 | 0.13 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr5_-_137374288 | 0.13 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr16_+_2479390 | 0.13 |
ENST00000397066.4
|
CCNF
|
cyclin F |
| chr14_+_104182061 | 0.13 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr6_-_135271260 | 0.12 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr4_-_174255400 | 0.12 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr14_+_57671888 | 0.12 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr8_+_22132847 | 0.11 |
ENST00000521356.1
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr3_-_105587879 | 0.11 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr8_+_22132810 | 0.10 |
ENST00000356766.6
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr6_+_160542870 | 0.10 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr17_+_36508826 | 0.10 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chrX_-_21676442 | 0.10 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr21_+_35014829 | 0.10 |
ENST00000451686.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr5_-_159546396 | 0.10 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr15_-_58571445 | 0.10 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr4_+_108911036 | 0.10 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr7_+_142985467 | 0.10 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr3_-_160823040 | 0.10 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr2_-_227050079 | 0.09 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr22_+_24322322 | 0.09 |
ENST00000215780.5
ENST00000402588.3 |
GSTT2
|
glutathione S-transferase theta 2 |
| chr14_+_32798547 | 0.09 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_209602156 | 0.09 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr1_+_87595433 | 0.09 |
ENST00000469312.2
ENST00000490006.2 |
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr2_+_135809835 | 0.08 |
ENST00000264158.8
ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr16_-_3493528 | 0.08 |
ENST00000301744.4
|
ZNF597
|
zinc finger protein 597 |
| chr5_-_159846399 | 0.08 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr1_-_23670813 | 0.07 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr13_+_111748183 | 0.07 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr9_+_104296243 | 0.07 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr1_-_68698197 | 0.07 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr10_-_99447024 | 0.07 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr19_+_1275917 | 0.06 |
ENST00000469144.1
|
C19orf24
|
chromosome 19 open reading frame 24 |
| chr11_-_124981475 | 0.06 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr8_-_117886612 | 0.06 |
ENST00000520992.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr10_+_91461413 | 0.06 |
ENST00000447580.1
|
KIF20B
|
kinesin family member 20B |
| chr17_-_74733404 | 0.06 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr16_+_3333443 | 0.06 |
ENST00000572748.1
ENST00000573578.1 ENST00000574253.1 |
ZNF263
|
zinc finger protein 263 |
| chr8_+_21881636 | 0.06 |
ENST00000520125.1
ENST00000521157.1 ENST00000397940.1 ENST00000522813.1 |
NPM2
|
nucleophosmin/nucleoplasmin 2 |
| chr2_+_242089833 | 0.05 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr1_+_40713573 | 0.05 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr11_-_118023490 | 0.05 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr14_-_24711764 | 0.05 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr2_-_198540719 | 0.05 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr3_+_111393501 | 0.05 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr22_+_17956618 | 0.05 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr10_-_1095050 | 0.05 |
ENST00000381344.3
|
IDI1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr11_-_111741994 | 0.05 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr4_+_40201954 | 0.04 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr6_-_10115007 | 0.04 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr6_+_36164487 | 0.04 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr14_-_73924954 | 0.04 |
ENST00000555307.1
ENST00000554818.1 |
NUMB
|
numb homolog (Drosophila) |
| chr6_-_30080876 | 0.04 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr1_+_220267429 | 0.04 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr14_+_91591873 | 0.04 |
ENST00000521064.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr22_-_24303340 | 0.04 |
ENST00000404172.3
ENST00000290765.4 |
GSTT2B
|
glutathione S-transferase theta 2B (gene/pseudogene) |
| chr20_+_9494987 | 0.04 |
ENST00000427562.2
ENST00000246070.2 |
LAMP5
|
lysosomal-associated membrane protein family, member 5 |
| chr3_-_113775328 | 0.04 |
ENST00000483766.1
ENST00000545063.1 ENST00000491000.1 ENST00000295878.3 |
KIAA1407
|
KIAA1407 |
| chr1_-_23670752 | 0.04 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr8_+_9413410 | 0.04 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr1_-_36916011 | 0.03 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr1_+_151735431 | 0.03 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.4 | 1.5 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.3 | 0.9 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.6 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.2 | 0.6 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 1.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 2.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.2 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 1.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.5 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.5 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 1.2 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.3 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 1.1 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 1.4 | GO:0006505 | GPI anchor metabolic process(GO:0006505) GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0035234 | luteolysis(GO:0001554) ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 1.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0006568 | tryptophan metabolic process(GO:0006568) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 1.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.4 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.1 | 1.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 0.6 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.2 | 0.6 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.2 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.6 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.5 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.4 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 0.5 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.2 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.1 | 1.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |