Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
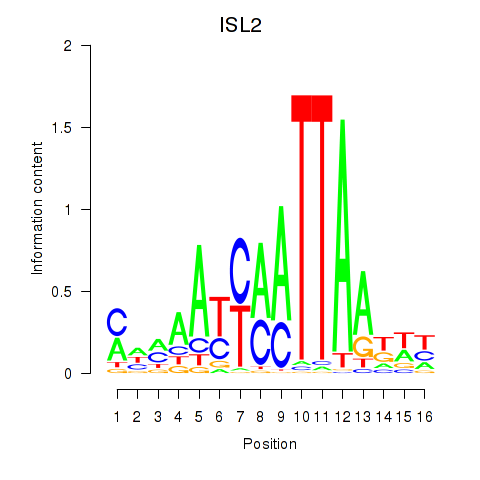
Results for ISL2
Z-value: 0.85
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.5 | ISL LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL2 | hg19_v2_chr15_+_76629064_76629073 | 0.96 | 3.7e-02 | Click! |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_327537 | 1.30 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr4_+_169013666 | 0.96 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr19_-_3557570 | 0.78 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr5_-_139726181 | 0.67 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr14_-_67878917 | 0.67 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr12_-_58212487 | 0.57 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr15_+_90735145 | 0.57 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr12_+_14369524 | 0.55 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr10_-_128359074 | 0.55 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr10_+_35484793 | 0.53 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr9_-_100854838 | 0.49 |
ENST00000538344.1
|
TRIM14
|
tripartite motif containing 14 |
| chr1_+_152178320 | 0.49 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr1_-_190446759 | 0.45 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_120243545 | 0.42 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr6_+_151561085 | 0.41 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr5_-_173217931 | 0.40 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr19_+_4402659 | 0.39 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_-_231860596 | 0.38 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr2_-_87248975 | 0.37 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr14_-_73997901 | 0.36 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr12_+_64798826 | 0.36 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr13_+_78315348 | 0.35 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr1_+_151253991 | 0.35 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr1_-_228613026 | 0.35 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr12_-_51422017 | 0.34 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr2_+_234826016 | 0.34 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr3_-_141747950 | 0.32 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_-_90756769 | 0.31 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_107729287 | 0.31 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_-_74800799 | 0.31 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr4_-_19458597 | 0.30 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr12_+_75784850 | 0.30 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chrX_-_55208866 | 0.28 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr2_-_136633940 | 0.27 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr10_-_95241951 | 0.27 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_-_121740969 | 0.27 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr12_-_118628315 | 0.26 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr11_-_62521614 | 0.26 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr13_-_46716969 | 0.26 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_-_183145873 | 0.26 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr2_-_158295915 | 0.26 |
ENST00000418920.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr5_+_40841276 | 0.25 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_-_178840157 | 0.25 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr6_+_42989344 | 0.25 |
ENST00000244496.5
|
RRP36
|
ribosomal RNA processing 36 homolog (S. cerevisiae) |
| chr18_-_64271316 | 0.24 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr10_-_95242044 | 0.24 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr19_+_14693888 | 0.24 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr2_-_217559517 | 0.23 |
ENST00000449583.1
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr20_-_33735070 | 0.23 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr19_+_3185910 | 0.23 |
ENST00000588428.1
|
NCLN
|
nicalin |
| chr4_+_26344754 | 0.23 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_107729504 | 0.22 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr6_+_31105426 | 0.22 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chrX_-_100872911 | 0.21 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr11_-_62609281 | 0.21 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr17_-_47786375 | 0.20 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr11_-_107729887 | 0.20 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr13_+_49551020 | 0.20 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr12_-_10282836 | 0.19 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chrX_-_84634708 | 0.19 |
ENST00000373145.3
|
POF1B
|
premature ovarian failure, 1B |
| chr19_-_46234119 | 0.19 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chrX_-_77225135 | 0.18 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr13_+_24144796 | 0.18 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr14_-_23058063 | 0.18 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr14_+_92789498 | 0.18 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr12_+_122356488 | 0.17 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr1_-_168464875 | 0.17 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr5_+_102200948 | 0.17 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_+_64680003 | 0.17 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr2_+_105050794 | 0.17 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr4_-_69111401 | 0.17 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr2_-_217560248 | 0.16 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr2_+_234668894 | 0.16 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr20_+_30102231 | 0.15 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr1_-_21377383 | 0.15 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr17_-_48785216 | 0.15 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr7_-_45151272 | 0.15 |
ENST00000461363.1
ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chr14_+_61449197 | 0.15 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr2_-_145277569 | 0.15 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_25162253 | 0.15 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr12_-_46121554 | 0.15 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr15_+_48483736 | 0.13 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr1_-_13390765 | 0.13 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr14_-_90798273 | 0.13 |
ENST00000357904.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr2_+_88047606 | 0.12 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr11_+_101983176 | 0.12 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr14_-_60043524 | 0.12 |
ENST00000537690.2
ENST00000281581.4 |
CCDC175
|
coiled-coil domain containing 175 |
| chr15_+_79166065 | 0.12 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chrX_-_48931648 | 0.12 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr6_-_47445214 | 0.11 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr1_-_232651312 | 0.11 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr18_-_3845293 | 0.11 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chrX_-_15332665 | 0.11 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr3_-_157221380 | 0.11 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr5_-_130500922 | 0.11 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr2_-_169887827 | 0.11 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr9_-_99540328 | 0.11 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr11_-_63376013 | 0.10 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chrX_-_32173579 | 0.09 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr7_-_107883678 | 0.09 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr3_+_169629354 | 0.09 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr17_+_7155819 | 0.09 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr18_-_64271363 | 0.09 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr1_-_95391315 | 0.09 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr1_+_15668240 | 0.09 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr5_-_54988559 | 0.09 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr12_-_118628350 | 0.08 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr1_+_160370344 | 0.08 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr7_-_55930443 | 0.08 |
ENST00000388975.3
|
SEPT14
|
septin 14 |
| chr4_+_26324474 | 0.08 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_-_166060382 | 0.08 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr10_-_74283694 | 0.08 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr1_-_223853348 | 0.08 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr9_-_28670283 | 0.08 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr5_+_40909354 | 0.07 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr1_+_149871135 | 0.07 |
ENST00000369152.5
|
BOLA1
|
bolA family member 1 |
| chr1_+_63063152 | 0.07 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chrX_+_102840408 | 0.07 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr14_+_38677123 | 0.07 |
ENST00000267377.2
|
SSTR1
|
somatostatin receptor 1 |
| chr3_+_159943362 | 0.07 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_-_152639479 | 0.07 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_41794313 | 0.06 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr14_-_90798418 | 0.06 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr18_-_53303123 | 0.06 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr15_-_42500351 | 0.06 |
ENST00000348544.4
ENST00000318006.5 |
VPS39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr8_+_92261516 | 0.06 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr3_+_134514093 | 0.06 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr8_+_38662960 | 0.06 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr21_+_39644305 | 0.06 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_85108363 | 0.06 |
ENST00000335459.5
|
TRABD2A
|
TraB domain containing 2A |
| chr11_+_12132117 | 0.06 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chrX_+_65384182 | 0.06 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr20_-_50722183 | 0.05 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr19_-_36001113 | 0.05 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr10_-_29923893 | 0.05 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr10_-_5046042 | 0.05 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr8_-_116504448 | 0.05 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr12_-_10282742 | 0.05 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_+_73498898 | 0.05 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr12_-_54689532 | 0.05 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr7_-_151330218 | 0.05 |
ENST00000476632.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_+_65382433 | 0.04 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr4_-_48116540 | 0.04 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr18_-_3845321 | 0.04 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr17_-_7307358 | 0.04 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr4_+_169418255 | 0.04 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_-_115670804 | 0.04 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr2_+_74685413 | 0.04 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr2_+_32502952 | 0.03 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr9_-_95166841 | 0.03 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr21_+_39644395 | 0.02 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr13_+_24144509 | 0.02 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr8_-_30706608 | 0.02 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chrX_+_65382381 | 0.02 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr7_-_115670792 | 0.02 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr11_+_86085778 | 0.02 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr9_-_21482312 | 0.01 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr1_+_149871171 | 0.01 |
ENST00000369150.1
|
BOLA1
|
bolA family member 1 |
| chr2_-_190446738 | 0.01 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr7_+_13141097 | 0.01 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr12_-_95945246 | 0.01 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chrM_+_9207 | 0.00 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_+_4710391 | 0.00 |
ENST00000263088.6
ENST00000572940.1 |
PLD2
|
phospholipase D2 |
| chr2_-_197675000 | 0.00 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr1_-_238108575 | 0.00 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr12_+_26164645 | 0.00 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.4 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.3 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 0.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 1.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.4 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |