Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ISL1
Z-value: 0.93
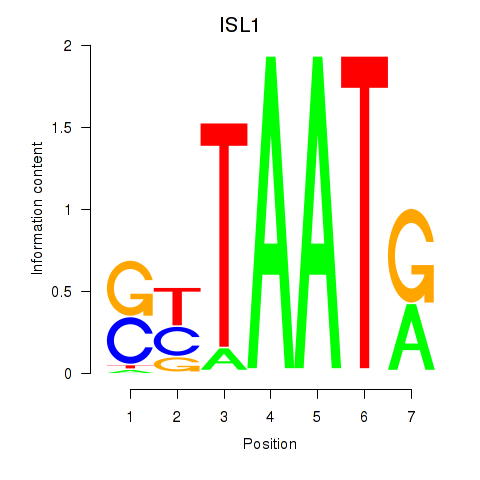
Transcription factors associated with ISL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL1
|
ENSG00000016082.10 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL1 | hg19_v2_chr5_+_50678921_50678921 | 0.77 | 2.3e-01 | Click! |
Activity profile of ISL1 motif
Sorted Z-values of ISL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_67957878 | 0.50 |
ENST00000420427.1
|
AC004562.1
|
AC004562.1 |
| chr6_+_116575329 | 0.33 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr20_-_5426380 | 0.32 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr14_+_22458631 | 0.30 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr13_+_28527647 | 0.27 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr5_+_140593509 | 0.27 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr12_-_94673956 | 0.26 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr12_+_14572070 | 0.26 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr20_+_60174827 | 0.24 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr19_+_39687596 | 0.24 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr2_-_175202151 | 0.24 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr18_+_21529811 | 0.22 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr4_+_74269956 | 0.22 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr2_+_207804278 | 0.22 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr11_-_46113756 | 0.21 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr8_-_131399110 | 0.21 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr14_-_21566731 | 0.21 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr11_-_111649015 | 0.21 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_-_102591604 | 0.21 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr12_-_42631529 | 0.20 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr3_-_186262166 | 0.20 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr19_+_3178736 | 0.19 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr3_+_157154578 | 0.19 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr7_-_122342988 | 0.19 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr5_+_137203557 | 0.18 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr21_+_17792672 | 0.18 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_220034745 | 0.17 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr3_-_179322436 | 0.16 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr14_+_62462541 | 0.16 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr4_+_139694701 | 0.16 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr11_-_7904464 | 0.16 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr7_+_13141010 | 0.16 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr12_-_57037284 | 0.15 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr9_+_131084846 | 0.15 |
ENST00000608951.1
|
COQ4
|
coenzyme Q4 |
| chr19_-_49622348 | 0.15 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr14_+_21566980 | 0.15 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr5_+_137203541 | 0.15 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr17_-_64225508 | 0.15 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr17_-_39023462 | 0.14 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr21_-_47575481 | 0.14 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr1_+_104159999 | 0.14 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr3_-_179322416 | 0.14 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr20_+_36405665 | 0.14 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr22_-_31885727 | 0.14 |
ENST00000330125.5
ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr12_+_1099675 | 0.14 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr4_-_168155169 | 0.14 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_-_94417314 | 0.14 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr21_-_19858196 | 0.13 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chrX_-_110655306 | 0.13 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr10_+_114710211 | 0.13 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr20_-_60294804 | 0.13 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr5_+_137203465 | 0.13 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr18_-_13915530 | 0.13 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr3_-_143567262 | 0.13 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr12_-_30887948 | 0.13 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr4_+_118955500 | 0.12 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_+_64018955 | 0.12 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr14_-_83262540 | 0.12 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr18_+_616672 | 0.12 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr6_+_31515337 | 0.12 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr5_-_102455801 | 0.11 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr1_+_197170592 | 0.11 |
ENST00000535699.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr12_+_21525818 | 0.11 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chrX_-_92928557 | 0.11 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr5_-_94417562 | 0.11 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_+_100598691 | 0.10 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr7_+_129932974 | 0.10 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr3_+_121774202 | 0.10 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr2_+_149402009 | 0.10 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr4_-_186877502 | 0.10 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_217804661 | 0.10 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr2_+_99758161 | 0.10 |
ENST00000409684.1
|
C2ORF15
|
Uncharacterized protein C2orf15 |
| chr4_+_4861385 | 0.10 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr16_-_4665023 | 0.10 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr13_+_78315348 | 0.10 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr8_+_19536083 | 0.10 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr2_+_170366203 | 0.10 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr2_+_202937972 | 0.10 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr9_-_96108696 | 0.10 |
ENST00000375419.1
|
C9orf129
|
chromosome 9 open reading frame 129 |
| chr3_-_47023455 | 0.09 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr2_+_234216454 | 0.09 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr9_+_128510454 | 0.09 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr4_-_186877806 | 0.09 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_+_103947311 | 0.09 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr17_-_7760457 | 0.09 |
ENST00000576384.1
|
LSMD1
|
LSM domain containing 1 |
| chr10_-_75118611 | 0.09 |
ENST00000355577.3
ENST00000394865.1 ENST00000310715.3 ENST00000401621.2 |
TTC18
|
tetratricopeptide repeat domain 18 |
| chr4_-_176923483 | 0.09 |
ENST00000280187.7
ENST00000512509.1 |
GPM6A
|
glycoprotein M6A |
| chr17_+_6918354 | 0.09 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr6_-_111136299 | 0.08 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr2_-_202563414 | 0.08 |
ENST00000409474.3
ENST00000315506.7 ENST00000359962.5 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr7_-_123673471 | 0.08 |
ENST00000455783.1
|
TMEM229A
|
transmembrane protein 229A |
| chr15_-_100258029 | 0.08 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr17_-_47045949 | 0.08 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr1_-_8000872 | 0.08 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr12_+_79357815 | 0.08 |
ENST00000547046.1
|
SYT1
|
synaptotagmin I |
| chr10_+_114710425 | 0.08 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_116681686 | 0.08 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_-_61190754 | 0.08 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr2_-_68547061 | 0.08 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr4_+_8201091 | 0.08 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr12_+_54402790 | 0.08 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr1_+_156024552 | 0.08 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr3_+_4345287 | 0.08 |
ENST00000358950.4
|
SETMAR
|
SET domain and mariner transposase fusion gene |
| chr1_+_223101757 | 0.08 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr16_-_67517716 | 0.08 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr1_+_100598742 | 0.08 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr2_+_172309634 | 0.08 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr2_-_158345341 | 0.08 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr3_-_27498235 | 0.08 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr12_+_54410664 | 0.08 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chr7_-_78400598 | 0.08 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr4_-_72649763 | 0.08 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr7_+_16793160 | 0.08 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr11_-_111649074 | 0.08 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_-_10251603 | 0.07 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr6_+_35996859 | 0.07 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr2_-_224467093 | 0.07 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_-_154842741 | 0.07 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr18_-_22804637 | 0.07 |
ENST00000577775.1
|
ZNF521
|
zinc finger protein 521 |
| chr8_-_1922789 | 0.07 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr1_+_15986364 | 0.07 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chrX_-_30327495 | 0.07 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chrX_-_138724994 | 0.07 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr12_-_10251539 | 0.07 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chrX_-_23926004 | 0.07 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr22_-_31885514 | 0.07 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr1_+_221054584 | 0.07 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr7_+_150811705 | 0.07 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr2_-_217560248 | 0.06 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr3_-_100551141 | 0.06 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_-_27529486 | 0.06 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr17_-_40729681 | 0.06 |
ENST00000590760.1
ENST00000587209.1 ENST00000393795.3 ENST00000253789.5 |
PSMC3IP
|
PSMC3 interacting protein |
| chrX_-_117107542 | 0.06 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr3_-_100712292 | 0.06 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_-_100712352 | 0.06 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr12_+_93096759 | 0.06 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr1_+_156024525 | 0.06 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr5_+_142286887 | 0.06 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr1_-_9953295 | 0.06 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr12_+_93096619 | 0.06 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr1_-_156217875 | 0.06 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr1_-_156217822 | 0.06 |
ENST00000368270.1
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr20_+_42136308 | 0.06 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr8_-_25281747 | 0.06 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr16_-_4664860 | 0.06 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr12_-_10251576 | 0.06 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr14_-_21567009 | 0.06 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr2_-_232329186 | 0.06 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chr12_-_28124903 | 0.06 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr5_-_41794313 | 0.06 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr15_+_48498480 | 0.05 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr7_+_138915102 | 0.05 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr4_+_169418255 | 0.05 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_+_113217043 | 0.05 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr18_+_616711 | 0.05 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr14_-_60337684 | 0.05 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr2_+_128377550 | 0.05 |
ENST00000437387.1
ENST00000409090.1 |
MYO7B
|
myosin VIIB |
| chr4_-_87281224 | 0.05 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr9_-_113761720 | 0.05 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chrX_-_117107680 | 0.05 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr2_+_219472637 | 0.05 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr8_-_116680833 | 0.05 |
ENST00000220888.5
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr17_-_39341594 | 0.05 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr4_+_68424434 | 0.05 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr12_-_53045948 | 0.05 |
ENST00000309680.3
|
KRT2
|
keratin 2 |
| chr11_+_22688150 | 0.05 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr20_-_48732472 | 0.05 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr19_+_13842559 | 0.05 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr9_+_37667978 | 0.05 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr8_-_133637624 | 0.05 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr17_-_80797886 | 0.05 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chrX_+_65382433 | 0.05 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr3_-_21792838 | 0.04 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr18_+_55018044 | 0.04 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr6_+_31707725 | 0.04 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr3_-_150966902 | 0.04 |
ENST00000424796.2
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr16_-_3350614 | 0.04 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chrX_+_65382381 | 0.04 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr4_+_159727272 | 0.04 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr3_-_146262637 | 0.04 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr3_+_130279178 | 0.04 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr11_-_118305921 | 0.04 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr12_+_9066472 | 0.04 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr6_+_90272027 | 0.04 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chrX_+_55478538 | 0.04 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr14_-_45252031 | 0.04 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr13_+_78315295 | 0.04 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_-_227050079 | 0.04 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr22_-_32651326 | 0.04 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr1_+_197382957 | 0.04 |
ENST00000367397.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr11_-_66496430 | 0.04 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr3_-_62359180 | 0.04 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chrX_-_15619076 | 0.03 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr11_+_120973375 | 0.03 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr4_-_87281196 | 0.03 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr10_-_62332357 | 0.03 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr19_-_50143452 | 0.03 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr2_-_214016314 | 0.03 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr13_-_95131923 | 0.03 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr6_-_123958141 | 0.03 |
ENST00000334268.4
|
TRDN
|
triadin |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |