Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
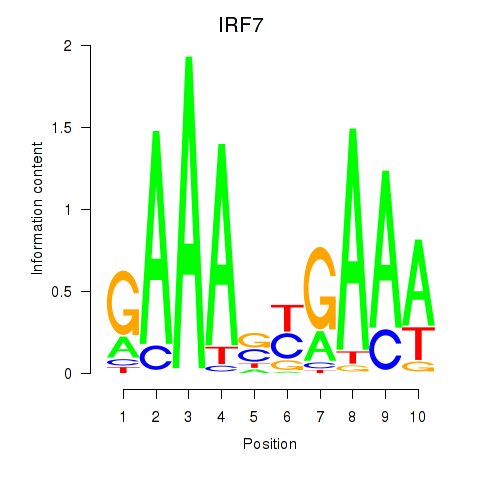
Results for IRF7
Z-value: 1.18
Transcription factors associated with IRF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF7
|
ENSG00000185507.15 | interferon regulatory factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF7 | hg19_v2_chr11_-_615942_615999 | 0.10 | 9.0e-01 | Click! |
Activity profile of IRF7 motif
Sorted Z-values of IRF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_91152303 | 0.70 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr4_-_169401628 | 0.65 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr12_+_113416265 | 0.60 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr1_-_150693318 | 0.58 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr10_-_9801179 | 0.55 |
ENST00000419836.1
|
RP5-1051H14.2
|
RP5-1051H14.2 |
| chr3_-_114035026 | 0.54 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr6_-_85473073 | 0.49 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr1_-_150693305 | 0.47 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr4_-_14889791 | 0.45 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr19_-_39735646 | 0.44 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr2_-_152118276 | 0.44 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr10_+_91061712 | 0.42 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr10_-_5446786 | 0.42 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr1_+_155829341 | 0.41 |
ENST00000539162.1
|
SYT11
|
synaptotagmin XI |
| chr20_-_32580924 | 0.39 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr5_+_121297650 | 0.37 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr4_-_130014729 | 0.37 |
ENST00000281142.5
ENST00000434680.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr5_-_133706695 | 0.37 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr16_-_11350036 | 0.35 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr17_-_62499334 | 0.35 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr18_-_21977748 | 0.35 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr17_-_57229155 | 0.35 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr9_+_21409146 | 0.35 |
ENST00000380205.1
|
IFNA8
|
interferon, alpha 8 |
| chr14_-_55738788 | 0.35 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr7_-_111424506 | 0.34 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chrX_-_71525742 | 0.34 |
ENST00000450875.1
ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr7_-_76955563 | 0.34 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr2_-_118943930 | 0.32 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr2_+_99771418 | 0.32 |
ENST00000393473.2
ENST00000393477.3 ENST00000393474.3 ENST00000340066.1 ENST00000393471.2 ENST00000449211.1 ENST00000434566.1 ENST00000410042.1 |
LIPT1
MRPL30
|
lipoyltransferase 1 39S ribosomal protein L30, mitochondrial |
| chr8_+_42873548 | 0.32 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr2_-_128785619 | 0.31 |
ENST00000450957.1
|
SAP130
|
Sin3A-associated protein, 130kDa |
| chr1_-_115301235 | 0.31 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr11_+_17298543 | 0.30 |
ENST00000533926.1
|
NUCB2
|
nucleobindin 2 |
| chr15_+_63354769 | 0.30 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr1_+_79086088 | 0.30 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr11_+_17298522 | 0.30 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr17_+_15604513 | 0.30 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr8_+_9009296 | 0.30 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr7_-_92146729 | 0.29 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr6_+_31105426 | 0.29 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chrX_+_51486481 | 0.29 |
ENST00000340438.4
|
GSPT2
|
G1 to S phase transition 2 |
| chr21_+_35553045 | 0.29 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr2_-_55237484 | 0.29 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr5_+_131746575 | 0.29 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr21_+_34144411 | 0.28 |
ENST00000382375.4
ENST00000453404.1 ENST00000382378.1 ENST00000477513.1 |
C21orf49
|
chromosome 21 open reading frame 49 |
| chr1_+_104068562 | 0.28 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr11_+_129936836 | 0.28 |
ENST00000597197.1
|
AP003041.2
|
Uncharacterized protein |
| chr5_+_180682720 | 0.28 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr4_-_40517984 | 0.28 |
ENST00000381795.6
|
RBM47
|
RNA binding motif protein 47 |
| chr10_+_91092241 | 0.27 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr4_-_187112626 | 0.27 |
ENST00000596414.1
|
AC110771.1
|
Uncharacterized protein |
| chr1_+_158979680 | 0.27 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr15_+_78832747 | 0.27 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr12_+_19358192 | 0.27 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr10_+_91174486 | 0.27 |
ENST00000416601.1
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr2_-_188419078 | 0.27 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr7_-_111424462 | 0.27 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr5_+_133707252 | 0.26 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr12_-_10324716 | 0.26 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr3_+_100120441 | 0.26 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chrX_+_78003204 | 0.26 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr11_-_5537920 | 0.26 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr3_+_122283175 | 0.26 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr12_+_40618764 | 0.25 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr14_+_73563735 | 0.25 |
ENST00000532192.1
|
RBM25
|
RNA binding motif protein 25 |
| chr4_-_76944621 | 0.25 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_+_158979792 | 0.25 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_-_108743471 | 0.24 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr14_-_35183755 | 0.24 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr8_-_30002179 | 0.24 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chrX_-_20237059 | 0.24 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr12_+_113416191 | 0.24 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr6_+_87865262 | 0.24 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr10_+_86184676 | 0.24 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr1_+_104104379 | 0.24 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr4_-_129207942 | 0.24 |
ENST00000503588.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr7_-_92777606 | 0.24 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_+_3875757 | 0.24 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr10_+_114710516 | 0.24 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_158979686 | 0.24 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr9_+_4985016 | 0.23 |
ENST00000539801.1
|
JAK2
|
Janus kinase 2 |
| chr3_-_28390581 | 0.23 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr15_-_80263506 | 0.23 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr10_+_91087651 | 0.23 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr2_+_153191706 | 0.23 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr6_+_142468383 | 0.23 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr5_+_68788594 | 0.23 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr9_-_119162885 | 0.23 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr11_-_119249805 | 0.22 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr19_+_49977466 | 0.22 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr3_-_187455680 | 0.22 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr11_+_9406169 | 0.22 |
ENST00000379719.3
ENST00000527431.1 |
IPO7
|
importin 7 |
| chr21_+_18811205 | 0.22 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chr1_+_59486129 | 0.22 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr18_+_61575200 | 0.22 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr3_+_185046676 | 0.22 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr11_-_60720002 | 0.22 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr12_+_19358228 | 0.21 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr1_+_104068312 | 0.21 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr12_+_113416340 | 0.21 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chrX_-_71526999 | 0.21 |
ENST00000453707.2
ENST00000373619.3 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr11_-_47736896 | 0.21 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr6_+_134273300 | 0.21 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr6_-_35888905 | 0.21 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr14_+_32546145 | 0.20 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr8_+_72755367 | 0.20 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr5_+_38845960 | 0.20 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr15_+_96875657 | 0.20 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_38978847 | 0.20 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr2_+_172950227 | 0.20 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr1_+_159796534 | 0.20 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr17_+_6659153 | 0.20 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr1_+_186265399 | 0.20 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr13_+_20532848 | 0.20 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr6_+_122931366 | 0.20 |
ENST00000368452.2
ENST00000368448.1 ENST00000392490.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr9_+_67977438 | 0.20 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr2_-_191878874 | 0.20 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr2_-_37384175 | 0.20 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr18_+_52258390 | 0.20 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr4_-_39034542 | 0.19 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr10_-_92681033 | 0.19 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_+_150245177 | 0.19 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr17_-_40264321 | 0.19 |
ENST00000430773.1
ENST00000413196.2 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr19_-_58485895 | 0.19 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr14_-_54908043 | 0.19 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr22_-_22641542 | 0.19 |
ENST00000606654.1
|
LL22NC03-2H8.5
|
LL22NC03-2H8.5 |
| chr6_+_116575329 | 0.19 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr7_+_142498725 | 0.19 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr14_-_69263043 | 0.19 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr15_+_42565844 | 0.19 |
ENST00000566442.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr7_-_83824449 | 0.18 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_129208030 | 0.18 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr15_-_77712429 | 0.18 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr2_-_191878681 | 0.18 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr4_-_169239921 | 0.18 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr9_-_32526184 | 0.18 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr3_+_97483572 | 0.18 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr8_-_79717750 | 0.18 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr8_+_27632083 | 0.18 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr11_+_93861993 | 0.18 |
ENST00000227638.3
ENST00000436171.2 |
PANX1
|
pannexin 1 |
| chr10_+_22610876 | 0.18 |
ENST00000442508.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr1_-_114430169 | 0.18 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr4_+_187112674 | 0.17 |
ENST00000378802.4
|
CYP4V2
|
cytochrome P450, family 4, subfamily V, polypeptide 2 |
| chr1_-_207119738 | 0.17 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr7_+_112063192 | 0.17 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr20_-_56195449 | 0.17 |
ENST00000541799.1
|
ZBP1
|
Z-DNA binding protein 1 |
| chr1_-_89488510 | 0.17 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr5_-_146461027 | 0.17 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_+_72058130 | 0.17 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr18_-_72264805 | 0.17 |
ENST00000577806.1
|
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr5_-_78281623 | 0.17 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr3_-_28389922 | 0.17 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr9_-_20622478 | 0.17 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr8_-_101733794 | 0.17 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_+_40841410 | 0.17 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr11_-_18258342 | 0.16 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr5_+_133706865 | 0.16 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr5_+_38846101 | 0.16 |
ENST00000274276.3
|
OSMR
|
oncostatin M receptor |
| chr17_-_40264692 | 0.16 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr1_-_111506562 | 0.16 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr10_+_24498060 | 0.16 |
ENST00000376454.3
ENST00000376452.3 |
KIAA1217
|
KIAA1217 |
| chr6_+_147527103 | 0.16 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr17_-_39156138 | 0.16 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr9_-_123812542 | 0.16 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr2_-_231084659 | 0.16 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr6_+_26440700 | 0.16 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr12_-_31479045 | 0.16 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr13_-_86373536 | 0.16 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr17_-_65362678 | 0.16 |
ENST00000357146.4
ENST00000356126.3 |
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr14_+_90864504 | 0.16 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chrX_+_146993534 | 0.16 |
ENST00000334557.6
ENST00000439526.2 ENST00000370475.4 |
FMR1
|
fragile X mental retardation 1 |
| chr12_-_72057571 | 0.15 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr5_-_58571935 | 0.15 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr17_+_6659354 | 0.15 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr17_-_33905521 | 0.15 |
ENST00000225873.4
|
PEX12
|
peroxisomal biogenesis factor 12 |
| chr11_-_47869865 | 0.15 |
ENST00000530326.1
ENST00000532747.1 |
NUP160
|
nucleoporin 160kDa |
| chr3_+_122399444 | 0.15 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chrX_+_49969405 | 0.15 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr5_+_82373317 | 0.15 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr6_+_160693591 | 0.15 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr2_-_190044480 | 0.15 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_-_103746924 | 0.15 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_99873145 | 0.15 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr5_-_98262240 | 0.15 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr4_-_185395191 | 0.15 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr17_-_74489215 | 0.15 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr1_+_43613612 | 0.15 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr10_+_91174314 | 0.15 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr9_+_6215799 | 0.15 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr1_-_149783914 | 0.15 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr12_+_102271436 | 0.15 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr14_-_69262916 | 0.15 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr12_+_113376157 | 0.14 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr19_+_17516624 | 0.14 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr18_-_29264467 | 0.14 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr2_-_55277692 | 0.14 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr5_+_34915444 | 0.14 |
ENST00000336767.5
|
BRIX1
|
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr4_+_186990298 | 0.14 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr10_+_51549498 | 0.14 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr12_+_79258547 | 0.14 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
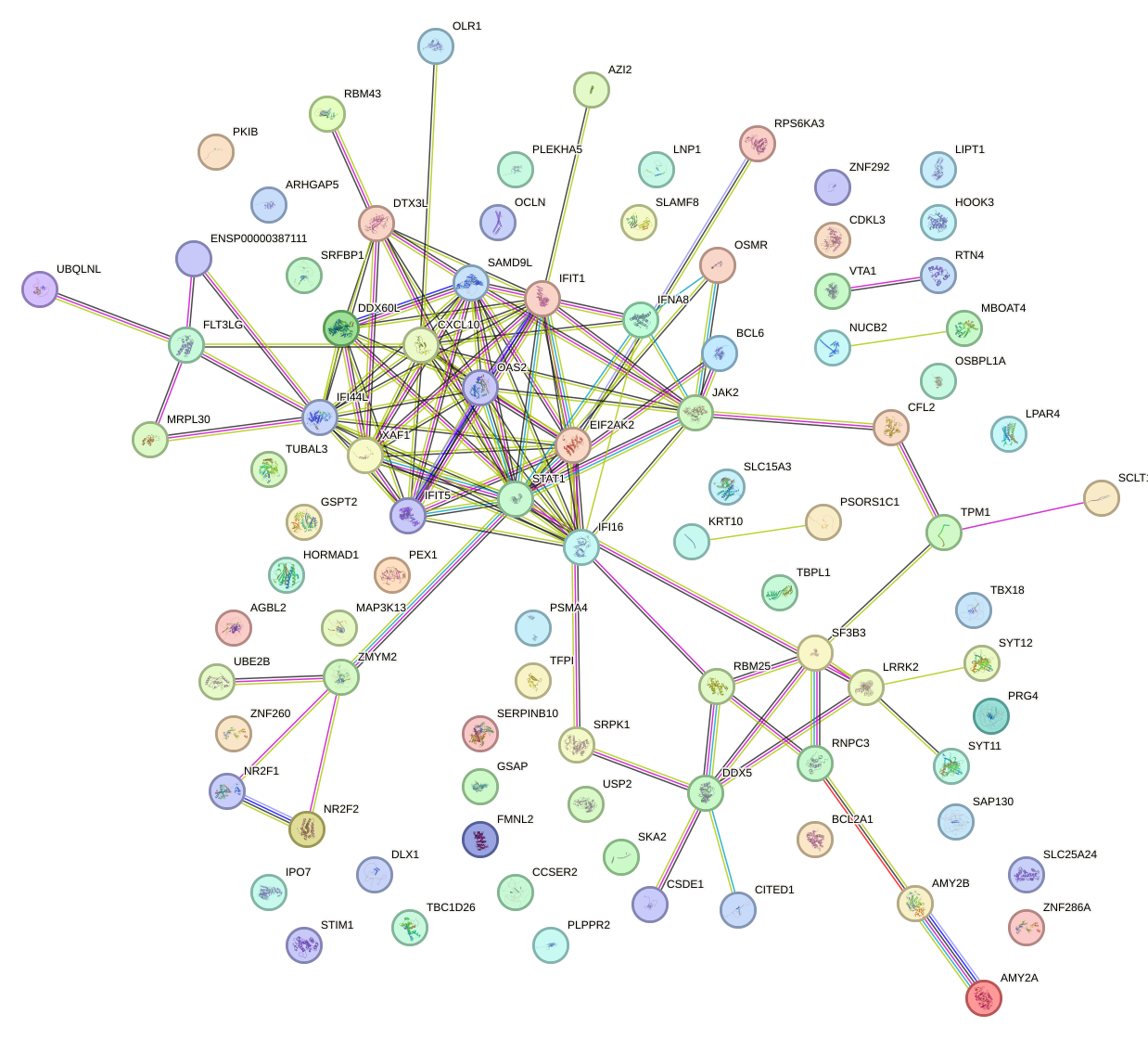
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0060629 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.2 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.6 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.5 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.4 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.5 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.0 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.3 | GO:1903217 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.4 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.2 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.6 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.2 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 0.2 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.4 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.2 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0090293 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.0 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0052056 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.0 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.4 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.0 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.0 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0045333 | cellular respiration(GO:0045333) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.2 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.0 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.4 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.3 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 0.4 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.8 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 2.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |