Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
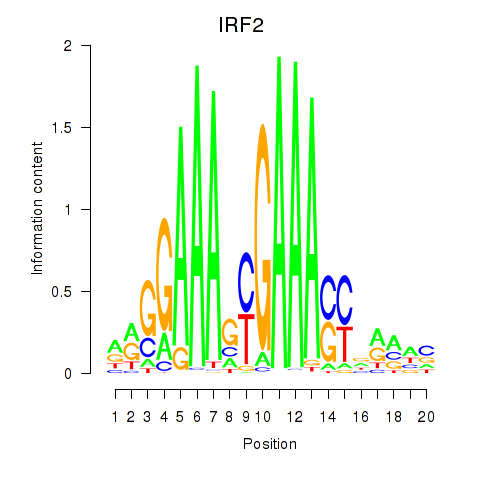
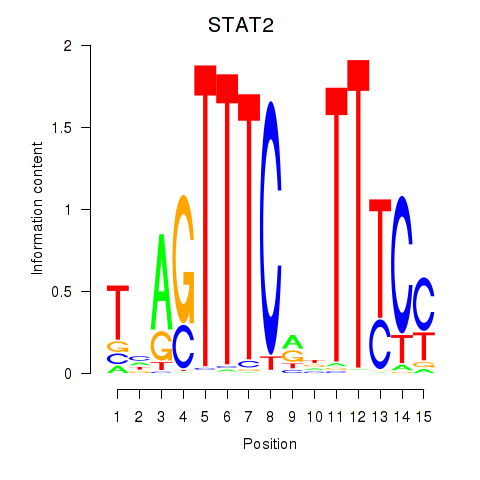
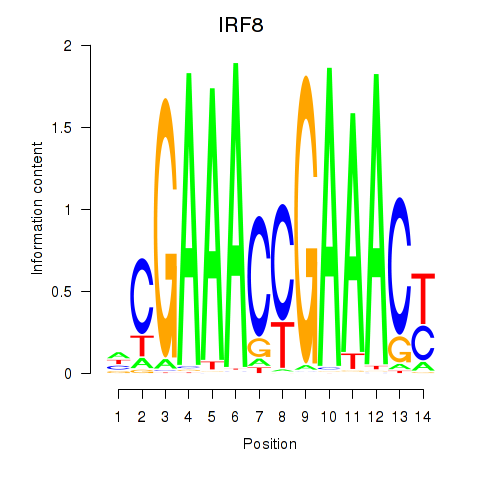
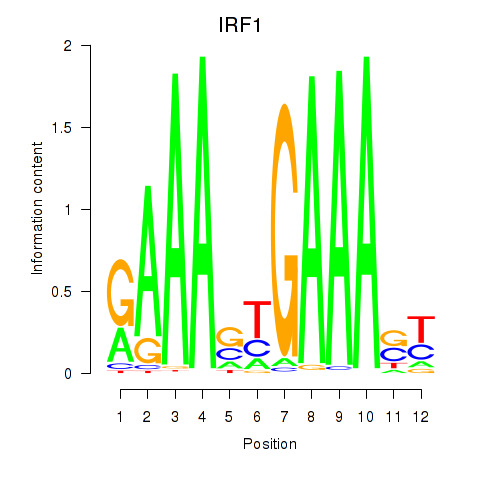
Results for IRF2_STAT2_IRF8_IRF1
Z-value: 16.83
Transcription factors associated with IRF2_STAT2_IRF8_IRF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF2
|
ENSG00000168310.6 | interferon regulatory factor 2 |
|
STAT2
|
ENSG00000170581.9 | signal transducer and activator of transcription 2 |
|
IRF8
|
ENSG00000140968.6 | interferon regulatory factor 8 |
|
IRF1
|
ENSG00000125347.9 | interferon regulatory factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT2 | hg19_v2_chr12_-_56753858_56753930 | 1.00 | 4.5e-03 | Click! |
| IRF1 | hg19_v2_chr5_-_131826457_131826514 | 0.99 | 8.7e-03 | Click! |
| IRF2 | hg19_v2_chr4_-_185395672_185395734 | 0.96 | 3.6e-02 | Click! |
| IRF8 | hg19_v2_chr16_+_85942594_85942635 | -0.59 | 4.1e-01 | Click! |
Activity profile of IRF2_STAT2_IRF8_IRF1 motif
Sorted Z-values of IRF2_STAT2_IRF8_IRF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_91152303 | 88.81 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr10_+_91087651 | 79.72 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr1_+_948803 | 77.51 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr10_+_91092241 | 72.82 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr12_+_113416191 | 67.42 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr2_-_7005785 | 59.02 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr1_+_79086088 | 56.54 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr21_+_42792442 | 54.02 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr22_+_18632666 | 53.31 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr11_-_64764435 | 51.17 |
ENST00000534177.1
ENST00000301887.4 |
BATF2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr10_+_91061712 | 50.05 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr11_-_615942 | 48.97 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr12_+_113416265 | 47.75 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr19_+_39759154 | 44.72 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr1_+_79115503 | 44.32 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr14_+_94577074 | 41.69 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr8_-_145060593 | 41.33 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr12_+_113376249 | 39.04 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr16_-_11350036 | 38.94 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr11_-_615570 | 37.85 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr6_+_32821924 | 33.37 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_+_7017796 | 33.28 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr17_+_6659153 | 32.84 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr16_+_57023406 | 31.35 |
ENST00000262510.6
ENST00000308149.7 ENST00000436936.1 |
NLRC5
|
NLR family, CARD domain containing 5 |
| chr7_-_92777606 | 31.25 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr12_+_113376157 | 30.68 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr2_-_231084820 | 30.52 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr9_-_32526299 | 29.85 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr2_-_163175133 | 29.44 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr2_+_7005959 | 29.13 |
ENST00000442639.1
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr3_-_49851313 | 29.02 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr3_+_187086120 | 28.87 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr1_+_158979680 | 28.69 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr4_+_89378261 | 28.56 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr2_-_231084659 | 28.53 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr2_-_191878874 | 28.24 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr2_-_191878681 | 28.05 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_158979686 | 27.70 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979792 | 27.62 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_-_27998689 | 27.60 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr4_-_169401628 | 27.29 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr9_-_32526184 | 26.64 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr21_+_42733870 | 25.90 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr11_+_5710919 | 25.52 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr3_-_122283424 | 24.12 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr1_-_89531041 | 23.85 |
ENST00000370473.4
|
GBP1
|
guanylate binding protein 1, interferon-inducible |
| chr9_-_100881466 | 23.81 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr3_-_122283100 | 23.68 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr21_+_42798094 | 23.40 |
ENST00000398598.3
ENST00000455164.2 ENST00000424365.1 |
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr6_-_32821599 | 23.34 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr10_+_91174486 | 22.90 |
ENST00000416601.1
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr3_-_122283079 | 22.64 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr10_+_91174314 | 22.19 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr19_-_49371711 | 22.14 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr2_-_55920952 | 21.75 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr19_+_10196781 | 21.47 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_+_5711010 | 21.09 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr11_-_4414880 | 21.03 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr9_+_100174232 | 20.43 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr9_+_100174344 | 20.27 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr12_+_113344811 | 19.83 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_-_39735646 | 19.82 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr22_-_36635684 | 19.75 |
ENST00000358502.5
|
APOL2
|
apolipoprotein L, 2 |
| chr9_+_5450503 | 19.72 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr6_+_32811885 | 19.59 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr22_+_36649056 | 19.53 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr11_-_321050 | 18.51 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr2_-_231084617 | 18.32 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr3_+_122399444 | 18.29 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr22_-_36635563 | 18.17 |
ENST00000451256.2
|
APOL2
|
apolipoprotein L, 2 |
| chr11_-_104905840 | 18.03 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr11_-_57334732 | 18.03 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr22_-_36635225 | 17.65 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr22_-_36635598 | 17.20 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr22_+_36649170 | 17.02 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr11_-_104916034 | 16.98 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr6_+_32811861 | 16.91 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr4_-_169239921 | 16.75 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr19_-_17516449 | 16.70 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr12_-_121477039 | 16.47 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr11_-_60719213 | 15.67 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr20_-_47894569 | 15.46 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr9_-_21077939 | 15.33 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr12_-_121476959 | 14.87 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_+_113416340 | 14.85 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr8_+_39770803 | 14.79 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr11_+_5646213 | 14.66 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr3_+_122283064 | 14.47 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr3_+_48507210 | 14.36 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr21_+_42798124 | 14.29 |
ENST00000417963.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr2_+_231280954 | 14.08 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr15_+_74287035 | 14.01 |
ENST00000395132.2
ENST00000268059.6 ENST00000354026.6 ENST00000268058.3 ENST00000565898.1 ENST00000569477.1 ENST00000569965.1 ENST00000567543.1 ENST00000436891.3 ENST00000435786.2 ENST00000564428.1 ENST00000359928.4 |
PML
|
promyelocytic leukemia |
| chr19_+_10196981 | 13.45 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr12_-_56753858 | 13.23 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr5_+_131746575 | 13.22 |
ENST00000337752.2
ENST00000378947.3 ENST00000407797.1 |
C5orf56
|
chromosome 5 open reading frame 56 |
| chr7_+_134832808 | 13.17 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr11_-_57335280 | 13.03 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr1_-_89488510 | 12.93 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr3_-_172241250 | 12.71 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr15_+_89182156 | 12.48 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_+_37400974 | 12.48 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr21_+_42798158 | 12.42 |
ENST00000441677.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr2_+_231280908 | 12.41 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr17_-_40264692 | 12.40 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr12_+_113344582 | 12.37 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_+_39786962 | 12.32 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr22_-_36556821 | 12.15 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr7_-_92747269 | 12.14 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chr15_+_89182178 | 12.13 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr22_+_39436862 | 12.08 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr21_+_42797958 | 12.01 |
ENST00000419044.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_+_69455855 | 11.62 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr8_-_23540402 | 11.44 |
ENST00000523261.1
ENST00000380871.4 |
NKX3-1
|
NK3 homeobox 1 |
| chr2_+_231191875 | 11.30 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr22_+_36044411 | 11.27 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr11_-_104972158 | 11.00 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr17_+_6659354 | 10.77 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr17_+_78234625 | 10.74 |
ENST00000508628.2
ENST00000582970.1 ENST00000456466.1 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr15_+_74287118 | 10.64 |
ENST00000563500.1
|
PML
|
promyelocytic leukemia |
| chr12_+_113344755 | 10.62 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr20_+_388791 | 10.62 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr11_-_63330842 | 10.41 |
ENST00000255695.1
|
HRASLS2
|
HRAS-like suppressor 2 |
| chr12_+_43086018 | 10.07 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr15_+_89181974 | 9.85 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr20_+_388935 | 9.80 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr11_+_71710973 | 9.76 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chr6_-_32806506 | 9.60 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr6_-_32811771 | 9.43 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_54991369 | 9.37 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chr2_-_37384175 | 9.06 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_+_48507621 | 9.02 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr20_+_388679 | 8.97 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr11_+_71710648 | 8.91 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr12_-_121476750 | 8.89 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr17_-_54991395 | 8.77 |
ENST00000316881.4
|
TRIM25
|
tripartite motif containing 25 |
| chr11_+_57308979 | 8.71 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr11_+_71709938 | 8.58 |
ENST00000393705.4
ENST00000337131.5 ENST00000531053.1 ENST00000404792.1 |
IL18BP
|
interleukin 18 binding protein |
| chr11_+_313503 | 8.55 |
ENST00000528780.1
ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr6_-_31324943 | 8.33 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr13_+_50070077 | 8.23 |
ENST00000378319.3
ENST00000426879.1 |
PHF11
|
PHD finger protein 11 |
| chr6_-_26027480 | 8.21 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr12_+_14369524 | 7.97 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr1_-_60392452 | 7.88 |
ENST00000371204.3
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2 |
| chr17_-_76975925 | 7.88 |
ENST00000591274.1
ENST00000589906.1 ENST00000591778.1 ENST00000589775.2 ENST00000585407.1 ENST00000262776.3 |
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr6_+_25963020 | 7.84 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr11_+_63304273 | 7.74 |
ENST00000439013.2
ENST00000255688.3 |
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3 |
| chr19_+_16254488 | 7.68 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr15_-_80263506 | 7.66 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr11_-_14358620 | 7.43 |
ENST00000531421.1
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr13_+_50070491 | 7.39 |
ENST00000496612.1
ENST00000357596.3 ENST00000485919.1 ENST00000442195.1 |
PHF11
|
PHD finger protein 11 |
| chr6_-_32806483 | 6.98 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_-_152146385 | 6.96 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr1_-_24469602 | 6.93 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr17_+_25958174 | 6.71 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr17_+_41158742 | 6.55 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr20_-_47894936 | 6.54 |
ENST00000371754.4
|
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr6_+_53659746 | 6.32 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr22_-_30695471 | 6.10 |
ENST00000434291.1
|
RP1-130H16.18
|
Uncharacterized protein |
| chr11_-_321340 | 5.98 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr1_+_113217309 | 5.92 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr3_-_146262365 | 5.90 |
ENST00000448787.2
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_+_113217345 | 5.78 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr12_-_54778471 | 5.73 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr3_-_146262352 | 5.73 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr12_-_54778444 | 5.70 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr12_-_10022735 | 5.69 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr14_+_101295948 | 5.66 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr1_+_113217073 | 5.63 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr3_-_178865747 | 5.52 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr3_-_146262428 | 5.50 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_+_113217043 | 5.47 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_-_43339474 | 5.43 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr6_+_26402465 | 5.38 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr3_-_146262637 | 5.30 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr6_+_29691056 | 5.25 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr3_+_122399697 | 5.14 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr6_+_29691198 | 5.08 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr22_-_36600727 | 5.04 |
ENST00000397275.2
ENST00000328429.4 |
APOL4
|
apolipoprotein L, 4 |
| chr3_-_28390415 | 5.02 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr17_-_43339453 | 4.97 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr3_+_122283175 | 4.88 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr17_+_7358889 | 4.85 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr4_-_185395672 | 4.79 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr6_+_126240442 | 4.77 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_-_170599561 | 4.71 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr1_-_151319654 | 4.68 |
ENST00000430227.1
ENST00000412774.1 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr9_-_21368075 | 4.68 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr17_-_4167142 | 4.60 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr15_+_74287009 | 4.33 |
ENST00000395135.3
|
PML
|
promyelocytic leukemia |
| chr3_-_146262488 | 4.28 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr11_-_47270341 | 4.27 |
ENST00000529444.1
ENST00000530453.1 ENST00000537863.1 ENST00000529788.1 ENST00000444355.2 ENST00000527256.1 ENST00000529663.1 ENST00000256997.3 |
ACP2
|
acid phosphatase 2, lysosomal |
| chr14_-_69445968 | 4.21 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr12_+_75874460 | 4.20 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr1_+_901847 | 4.17 |
ENST00000379410.3
ENST00000379409.2 ENST00000379407.3 |
PLEKHN1
|
pleckstrin homology domain containing, family N member 1 |
| chr4_+_185395947 | 4.17 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr7_+_150382781 | 4.07 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr2_+_210288760 | 4.05 |
ENST00000199940.6
|
MAP2
|
microtubule-associated protein 2 |
| chr21_+_43933946 | 4.03 |
ENST00000352133.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr7_-_134143841 | 4.00 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr1_-_154580616 | 3.96 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF2_STAT2_IRF8_IRF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.6 | 90.5 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 22.3 | 89.3 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 22.2 | 88.8 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 14.8 | 59.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 14.5 | 202.6 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 14.1 | 56.3 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 13.8 | 166.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 13.5 | 40.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 11.8 | 130.0 | GO:0018377 | protein myristoylation(GO:0018377) |
| 10.6 | 63.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 10.0 | 39.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 8.1 | 73.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 8.0 | 88.4 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 7.3 | 21.8 | GO:2000625 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 6.9 | 34.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 6.8 | 142.0 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 6.7 | 26.9 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 5.9 | 70.7 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 5.6 | 16.7 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 5.2 | 20.9 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 4.9 | 14.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 4.7 | 37.6 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 4.6 | 27.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 4.2 | 12.5 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 3.8 | 379.8 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 3.7 | 18.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 3.2 | 48.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 2.9 | 29.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 2.9 | 192.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 2.7 | 10.7 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 2.5 | 49.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 2.4 | 28.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 2.3 | 7.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 2.1 | 23.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 2.0 | 14.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 2.0 | 15.9 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 1.9 | 11.4 | GO:0060664 | androgen secretion(GO:0035935) epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 1.8 | 10.6 | GO:0030421 | defecation(GO:0030421) |
| 1.6 | 4.8 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 1.6 | 1.6 | GO:0061209 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) cell-cell signaling involved in cell fate commitment(GO:0045168) cell proliferation involved in mesonephros development(GO:0061209) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 1.6 | 4.7 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) positive regulation of skeletal muscle tissue growth(GO:0048633) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 1.5 | 40.1 | GO:0002385 | mucosal immune response(GO:0002385) |
| 1.4 | 2.8 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.3 | 4.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.3 | 1.3 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 1.2 | 12.3 | GO:0070141 | response to UV-A(GO:0070141) |
| 1.2 | 12.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 1.1 | 40.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 1.1 | 26.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 1.0 | 27.0 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 1.0 | 3.0 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.0 | 8.7 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 1.0 | 37.2 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.9 | 2.7 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.9 | 4.6 | GO:0060611 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.9 | 23.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.9 | 67.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.8 | 0.8 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.8 | 4.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.8 | 71.6 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.8 | 7.7 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.7 | 2.8 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.7 | 2.1 | GO:0070859 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.7 | 8.4 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.7 | 1.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 2.0 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.7 | 2.6 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.7 | 19.1 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.6 | 1.9 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.6 | 1.8 | GO:0072300 | vestibulocochlear nerve formation(GO:0021650) negative regulation of cytolysis(GO:0045918) positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.5 | 56.7 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.5 | 4.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.5 | 3.8 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 1.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.5 | 2.7 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.4 | 5.8 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.4 | 2.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 2.2 | GO:1904582 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.4 | 1.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.4 | 1.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.4 | 1.2 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.4 | 4.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.4 | 2.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.4 | 3.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.4 | 1.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 8.4 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.4 | 2.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 1.4 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.3 | 2.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.3 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 1.0 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.3 | 1.9 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 0.9 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.3 | 5.8 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.3 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 15.0 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.3 | 7.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.3 | 1.2 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.3 | 1.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 2.6 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.3 | 1.7 | GO:0051271 | negative regulation of cellular component movement(GO:0051271) |
| 0.3 | 0.3 | GO:1903383 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.3 | 1.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 2.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 7.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 1.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 2.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.3 | 7.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 0.5 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.2 | 0.9 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 1.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 0.2 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 1.7 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.2 | 13.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 0.6 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.2 | 0.4 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.2 | 1.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 1.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.2 | 1.6 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.2 | 1.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.5 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 0.5 | GO:0061074 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) regulation of neural retina development(GO:0061074) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 1.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.2 | GO:0061458 | reproductive system development(GO:0061458) |
| 0.2 | 2.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 1.5 | GO:0043473 | pigmentation(GO:0043473) |
| 0.2 | 0.3 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.2 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 1.3 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 1.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.2 | 2.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.2 | 0.6 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 0.8 | GO:0071609 | chemokine (C-C motif) ligand 5 production(GO:0071609) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.2 | 1.3 | GO:1901295 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 0.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 0.5 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 2.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.8 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.2 | 5.7 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.2 | 0.5 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 2.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.6 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.9 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 2.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 48.8 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 0.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 1.1 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.4 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 1.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) positive regulation of transcription by glucose(GO:0046016) |
| 0.1 | 0.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 4.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.9 | GO:0098704 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.3 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.5 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 3.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 2.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 1.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.9 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.9 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 | 3.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.7 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 0.6 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 2.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.5 | GO:0051174 | regulation of phosphate metabolic process(GO:0019220) regulation of phosphorus metabolic process(GO:0051174) |
| 0.1 | 0.6 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 1.8 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.3 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.4 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.5 | GO:0048839 | ear development(GO:0043583) inner ear development(GO:0048839) |
| 0.1 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.1 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.6 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.2 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.6 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.1 | 1.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.1 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.5 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.4 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 16.3 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 1.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.2 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 2.3 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 2.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 2.0 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.1 | 0.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.9 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.3 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.1 | 0.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 3.1 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.1 | 1.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.7 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.1 | 1.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.4 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.1 | 0.5 | GO:0002223 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 10.8 | GO:0006818 | hydrogen transport(GO:0006818) proton transport(GO:0015992) |
| 0.1 | 1.6 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 7.8 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 6.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.8 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.8 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.4 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.8 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 1.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 1.8 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.8 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.2 | GO:0042357 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 1.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.8 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 4.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.8 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.8 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.5 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.2 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.4 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.4 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) |
| 0.0 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.8 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 1.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 3.6 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.4 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 2.5 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.9 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.3 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0006024 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.7 | GO:0090109 | regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 60.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 6.6 | 46.0 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 5.9 | 41.2 | GO:0042825 | TAP complex(GO:0042825) |
| 4.9 | 29.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 4.1 | 29.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 4.1 | 12.3 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 4.0 | 12.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 2.3 | 40.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 2.1 | 49.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 1.9 | 24.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.8 | 24.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.3 | 7.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.2 | 36.1 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 1.0 | 14.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 1.0 | 80.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.9 | 6.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.8 | 21.6 | GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.8 | 4.6 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 2.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.7 | 4.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.7 | 4.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.6 | 2.4 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.6 | 4.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 1.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.6 | 68.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.6 | 70.5 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.5 | 4.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.4 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.4 | 74.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.4 | 1.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.4 | 9.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.4 | 20.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 2.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.3 | 1.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 1.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.3 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 33.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 0.9 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 1.4 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.3 | 4.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 0.8 | GO:0001534 | radial spoke(GO:0001534) |
| 0.3 | 9.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 106.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 49.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 0.7 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.2 | 15.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 1.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 14.8 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.2 | 4.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 1.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 7.8 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 31.5 | GO:0005769 | early endosome(GO:0005769) |
| 0.2 | 1.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 157.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 3.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 0.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 7.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 3.3 | GO:0036020 | endolysosome(GO:0036019) endolysosome membrane(GO:0036020) |
| 0.1 | 4.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 64.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 2.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 148.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 269.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 3.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 14.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 12.3 | GO:0031975 | organelle envelope(GO:0031967) envelope(GO:0031975) |
| 0.1 | 0.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 1.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 75.2 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.1 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.7 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.1 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 9.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.8 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 3.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 9.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 4.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 61.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 1.1 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 157.8 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.7 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 11.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 13.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.6 | 282.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 14.8 | 59.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 11.5 | 34.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 9.1 | 27.3 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 7.2 | 28.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 6.0 | 77.5 | GO:0031386 | protein tag(GO:0031386) |
| 6.0 | 59.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 5.7 | 51.5 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 4.2 | 25.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 4.2 | 12.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 4.1 | 73.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 3.7 | 15.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 3.5 | 24.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 3.1 | 15.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 2.9 | 26.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 2.9 | 29.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 2.9 | 17.3 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 2.9 | 137.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 2.2 | 28.0 | GO:0089720 | caspase binding(GO:0089720) |
| 2.0 | 59.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.9 | 9.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.9 | 11.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 1.7 | 6.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.5 | 8.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.4 | 44.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 1.4 | 69.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 1.3 | 29.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.1 | 6.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 1.1 | 6.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.1 | 2.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.0 | 40.9 | GO:0005521 | lamin binding(GO:0005521) |
| 1.0 | 126.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 1.0 | 62.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 1.0 | 18.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.0 | 3.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 1.0 | 7.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.8 | 2.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.8 | 18.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.7 | 3.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 96.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.7 | 36.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 18.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 1.9 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.6 | 1.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.6 | 1.2 | GO:0035877 | death effector domain binding(GO:0035877) cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.6 | 3.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 5.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.5 | 1.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.5 | 2.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.5 | 12.9 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.5 | 83.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.5 | 24.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.5 | 1.9 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.5 | 3.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 3.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.4 | 1.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 2.0 | GO:0050659 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.4 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 22.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 52.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 7.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.4 | 4.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.4 | 1.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.4 | 1.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.3 | 2.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 2.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.3 | 7.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 37.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.3 | 1.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 184.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.3 | 4.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 8.4 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.3 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.3 | 3.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 2.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 4.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 40.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 2.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 7.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 2.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.2 | 45.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 1.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.2 | 0.8 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 4.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 7.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.2 | 2.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.2 | 1.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 1.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 34.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.2 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.2 | 1.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.7 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 0.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.4 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 2.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 1.5 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 6.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 16.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 7.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.9 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 8.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 34.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.9 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 1.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 3.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.9 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 1.0 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.4 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 14.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 12.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 2.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 13.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.3 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 60.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 1.6 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.2 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.1 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 4.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 73.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 2.0 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 8.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 1.6 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.5 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 43.4 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 53.4 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 5.4 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 1.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.5 | GO:0003676 | nucleic acid binding(GO:0003676) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 88.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.8 | 28.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.7 | 11.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 33.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.5 | 38.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 23.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.5 | 14.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.4 | 70.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.4 | 40.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.4 | 23.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 17.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 14.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 19.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 2.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 12.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 13.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 28.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 6.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 13.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.5 | 1291.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 6.5 | 104.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 2.2 | 116.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 1.3 | 12.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.3 | 78.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 1.3 | 27.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.3 | 37.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.9 | 34.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.8 | 57.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.8 | 9.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.8 | 10.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.7 | 4.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 17.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 2.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 2.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 9.9 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.3 | 2.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.2 | 63.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 8.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.2 | 4.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 14.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 6.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 5.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 10.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 1.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 3.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.1 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |