Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
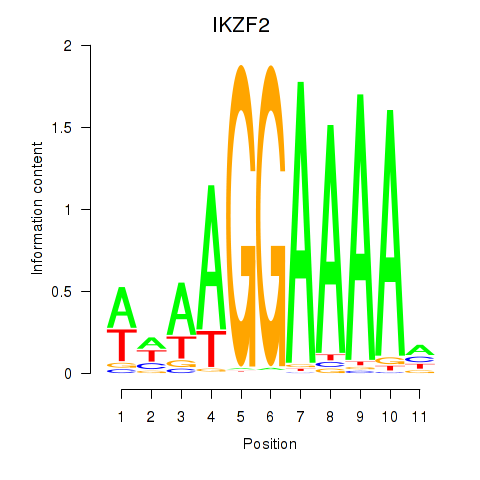
Results for IKZF2
Z-value: 0.68
Transcription factors associated with IKZF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IKZF2
|
ENSG00000030419.12 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | hg19_v2_chr2_-_214015111_214015179 | -0.72 | 2.8e-01 | Click! |
Activity profile of IKZF2 motif
Sorted Z-values of IKZF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_79115503 | 0.67 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr10_-_10504285 | 0.56 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr1_+_158979680 | 0.43 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979686 | 0.43 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr6_-_15586238 | 0.43 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr8_-_23282820 | 0.41 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr17_-_6915616 | 0.40 |
ENST00000575889.1
|
AC027763.2
|
Uncharacterized protein |
| chr3_+_156393349 | 0.39 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr15_+_45722727 | 0.35 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr8_-_123706338 | 0.35 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr12_+_15154767 | 0.34 |
ENST00000542689.1
|
RP11-508P1.2
|
RP11-508P1.2 |
| chr1_+_158979792 | 0.33 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr17_+_39405939 | 0.33 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr11_-_33891362 | 0.32 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_+_50433476 | 0.30 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr6_+_126221034 | 0.30 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr1_-_161337662 | 0.29 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr11_+_18417948 | 0.28 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr13_-_34250861 | 0.27 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr13_-_30160925 | 0.27 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_-_39735646 | 0.26 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr2_+_120517717 | 0.26 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr1_-_57045228 | 0.26 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr17_-_15469590 | 0.24 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr12_-_122240792 | 0.24 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr6_+_36839616 | 0.24 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr1_-_157014865 | 0.23 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr1_-_67600639 | 0.23 |
ENST00000544837.1
ENST00000603691.1 |
C1orf141
|
chromosome 1 open reading frame 141 |
| chr16_+_75033210 | 0.22 |
ENST00000566250.1
ENST00000567962.1 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr19_+_10197463 | 0.22 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_-_52931557 | 0.21 |
ENST00000504329.1
ENST00000355083.5 |
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr3_+_171844762 | 0.20 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr7_-_127671674 | 0.20 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr17_+_77019030 | 0.20 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr5_+_140180635 | 0.20 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr22_+_43011247 | 0.20 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr11_+_58938903 | 0.20 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr17_+_77018896 | 0.19 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_-_127963343 | 0.19 |
ENST00000335247.7
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr12_-_8693539 | 0.19 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr14_+_35591020 | 0.19 |
ENST00000603611.1
|
KIAA0391
|
KIAA0391 |
| chr12_-_8693469 | 0.18 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr17_-_39093672 | 0.18 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr20_+_58179582 | 0.18 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr11_+_126153001 | 0.18 |
ENST00000392678.3
ENST00000392680.2 |
TIRAP
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr22_-_39640756 | 0.18 |
ENST00000331163.6
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr7_+_66800928 | 0.18 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr17_+_35294075 | 0.17 |
ENST00000254457.5
|
LHX1
|
LIM homeobox 1 |
| chr11_+_18417813 | 0.17 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chrX_+_135252050 | 0.17 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr3_-_121740969 | 0.17 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr10_+_97759848 | 0.17 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr5_+_34685805 | 0.16 |
ENST00000508315.1
|
RAI14
|
retinoic acid induced 14 |
| chr6_+_149539053 | 0.16 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr9_-_16705069 | 0.16 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr12_+_9144626 | 0.16 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_+_104615595 | 0.16 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr10_+_115511434 | 0.16 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr7_-_127672146 | 0.16 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr3_-_178865747 | 0.16 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr1_-_47655686 | 0.16 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr2_+_33661382 | 0.15 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_73520667 | 0.15 |
ENST00000545030.1
ENST00000436467.2 |
EGR4
|
early growth response 4 |
| chr6_-_133055896 | 0.15 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr12_-_26278030 | 0.15 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chrX_+_135251835 | 0.15 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr14_+_91709279 | 0.15 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr12_-_104443890 | 0.15 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr14_+_35591928 | 0.15 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr11_+_47293795 | 0.15 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr14_+_23025534 | 0.15 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr2_-_190927447 | 0.15 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr14_+_63671105 | 0.15 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chrX_+_135251783 | 0.15 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr4_-_185303418 | 0.14 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr6_+_144665237 | 0.14 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr11_+_125464758 | 0.14 |
ENST00000529886.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr7_+_93551011 | 0.14 |
ENST00000248564.5
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr22_-_36220420 | 0.14 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_+_135230712 | 0.14 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr15_-_55541227 | 0.13 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr8_-_36636676 | 0.13 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr13_+_49551020 | 0.13 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr14_+_35591735 | 0.12 |
ENST00000604948.1
ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391
|
KIAA0391 |
| chr8_+_97597148 | 0.12 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr4_-_134070250 | 0.12 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr11_-_46142948 | 0.12 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chr20_+_42574317 | 0.12 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr15_-_82338460 | 0.12 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr19_+_18451391 | 0.12 |
ENST00000269919.6
ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr14_-_36988882 | 0.12 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr14_+_35591858 | 0.12 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chr15_-_59665062 | 0.11 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr11_-_19263145 | 0.11 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr9_+_19049372 | 0.11 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr15_+_66679155 | 0.11 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr2_+_101437487 | 0.11 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr17_+_57970469 | 0.11 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr1_+_230193521 | 0.11 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr13_-_46756351 | 0.11 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chrX_-_15683147 | 0.11 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr1_-_242612726 | 0.11 |
ENST00000459864.1
|
PLD5
|
phospholipase D family, member 5 |
| chr8_-_128231299 | 0.11 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr4_+_74347400 | 0.11 |
ENST00000226355.3
|
AFM
|
afamin |
| chr3_-_169487617 | 0.10 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr18_-_48351743 | 0.10 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr3_-_71294304 | 0.10 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr6_-_133055815 | 0.10 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr22_-_50699701 | 0.10 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr17_-_39211463 | 0.10 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr17_-_56082455 | 0.10 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr4_-_120243545 | 0.10 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr19_-_51141196 | 0.10 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr8_+_39770803 | 0.10 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr4_-_71532601 | 0.10 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_-_35591156 | 0.10 |
ENST00000554361.1
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr6_+_106534192 | 0.10 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr6_-_112081113 | 0.10 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr1_-_157015162 | 0.10 |
ENST00000368194.3
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr10_+_121578211 | 0.09 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr17_+_55055466 | 0.09 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr5_-_160973649 | 0.09 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr2_-_101767715 | 0.09 |
ENST00000376840.4
ENST00000409318.1 |
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain) |
| chr22_+_46449674 | 0.09 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr1_+_36038971 | 0.09 |
ENST00000373235.3
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
| chr6_-_11779014 | 0.09 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr17_+_27047244 | 0.09 |
ENST00000394938.4
ENST00000394935.3 ENST00000355731.4 |
RPL23A
|
ribosomal protein L23a |
| chr6_-_11779403 | 0.09 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr4_+_26322185 | 0.08 |
ENST00000361572.6
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_11779174 | 0.08 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_+_112938803 | 0.08 |
ENST00000271277.6
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr1_+_9711781 | 0.08 |
ENST00000536656.1
ENST00000377346.4 |
PIK3CD
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr6_+_116691001 | 0.08 |
ENST00000537543.1
|
DSE
|
dermatan sulfate epimerase |
| chr6_+_32944119 | 0.08 |
ENST00000606059.1
|
BRD2
|
bromodomain containing 2 |
| chrX_-_15619076 | 0.08 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr3_+_57882061 | 0.08 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr16_+_21623958 | 0.08 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr6_+_149539767 | 0.08 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chrX_+_48660287 | 0.08 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr8_-_102987548 | 0.08 |
ENST00000520690.1
|
NCALD
|
neurocalcin delta |
| chr12_-_80328905 | 0.08 |
ENST00000547330.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr15_-_66679019 | 0.08 |
ENST00000568216.1
ENST00000562124.1 ENST00000570251.1 |
TIPIN
|
TIMELESS interacting protein |
| chr17_+_27046988 | 0.08 |
ENST00000496182.1
|
RPL23A
|
ribosomal protein L23a |
| chr17_-_48785216 | 0.08 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr3_-_71632894 | 0.08 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr6_+_33388013 | 0.08 |
ENST00000449372.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chrX_+_48660107 | 0.08 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr17_-_17740325 | 0.07 |
ENST00000338854.5
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr17_+_6915730 | 0.07 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr16_-_73093597 | 0.07 |
ENST00000397992.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr4_+_130017268 | 0.07 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr7_-_111424506 | 0.07 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr10_-_79398250 | 0.07 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_-_6915646 | 0.07 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr10_-_79789291 | 0.07 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr12_-_118797475 | 0.07 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr19_-_41859814 | 0.07 |
ENST00000221930.5
|
TGFB1
|
transforming growth factor, beta 1 |
| chr17_+_7387677 | 0.07 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr9_-_73483926 | 0.07 |
ENST00000396283.1
ENST00000361823.5 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr19_+_18496957 | 0.07 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr12_-_49449107 | 0.07 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr9_+_22646189 | 0.07 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr8_-_82024290 | 0.07 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr14_-_45431091 | 0.07 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr12_+_103981044 | 0.07 |
ENST00000388887.2
|
STAB2
|
stabilin 2 |
| chr12_-_118796910 | 0.07 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr6_-_24721054 | 0.07 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr1_+_161719552 | 0.07 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr3_-_38992052 | 0.07 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr1_+_28995231 | 0.07 |
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr19_-_31840438 | 0.07 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr9_+_34989638 | 0.07 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr10_-_128975273 | 0.07 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chrX_-_30993201 | 0.07 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr10_+_48189612 | 0.07 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr4_-_74486347 | 0.07 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr8_-_19459993 | 0.06 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr21_+_26934165 | 0.06 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr1_-_68698222 | 0.06 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr7_-_122840015 | 0.06 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr1_+_2066387 | 0.06 |
ENST00000497183.1
|
PRKCZ
|
protein kinase C, zeta |
| chr18_-_47340297 | 0.06 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr9_-_91793675 | 0.06 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr9_-_130667592 | 0.06 |
ENST00000447681.1
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr16_+_20911722 | 0.06 |
ENST00000569023.1
|
LYRM1
|
LYR motif containing 1 |
| chr17_-_38928414 | 0.06 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr8_+_9183618 | 0.06 |
ENST00000518619.1
|
RP11-115J16.1
|
RP11-115J16.1 |
| chr8_-_145754428 | 0.06 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr19_+_11200038 | 0.06 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chrX_+_45364633 | 0.06 |
ENST00000435394.1
ENST00000609127.1 |
RP11-245M24.1
|
RP11-245M24.1 |
| chr7_-_149158187 | 0.06 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr9_-_15250114 | 0.06 |
ENST00000507993.1
|
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr17_-_57970074 | 0.06 |
ENST00000346141.6
|
TUBD1
|
tubulin, delta 1 |
| chr17_+_7387919 | 0.06 |
ENST00000572844.1
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr1_-_15850676 | 0.06 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr1_+_172502244 | 0.06 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr13_+_97874574 | 0.06 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr12_+_53835425 | 0.06 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr3_+_46618727 | 0.06 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr19_+_7599128 | 0.06 |
ENST00000545201.2
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IKZF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.2 | GO:2000338 | interleukin-15 production(GO:0032618) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.1 | 0.2 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.4 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:1990936 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.2 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.0 | GO:1903980 | positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.3 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0052255 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.6 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 1.2 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.1 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.4 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0097101 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |