Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
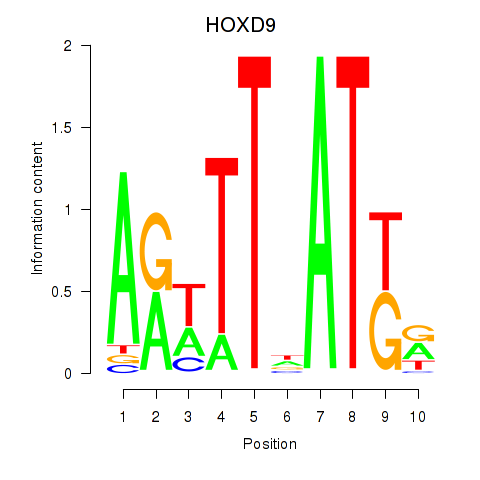
Results for HOXD9
Z-value: 0.57
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | 0.65 | 3.5e-01 | Click! |
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_92777606 | 0.38 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr5_+_68860949 | 0.29 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr13_-_41111323 | 0.28 |
ENST00000595486.1
|
AL133318.1
|
Uncharacterized protein |
| chr13_+_111748183 | 0.26 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr10_+_86184676 | 0.22 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr6_-_18249971 | 0.22 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr1_-_238108575 | 0.22 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr11_-_104905840 | 0.22 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr7_+_90012986 | 0.21 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr16_-_18887627 | 0.21 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_+_143635067 | 0.21 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr17_+_60447579 | 0.21 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr7_+_13141010 | 0.21 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr7_+_123469037 | 0.20 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr15_+_49913175 | 0.19 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr14_+_57671888 | 0.19 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr2_+_27282134 | 0.19 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr13_-_81801115 | 0.18 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr21_-_39705286 | 0.18 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chr8_+_101349823 | 0.18 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr5_-_159766528 | 0.17 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr7_-_130353553 | 0.17 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr12_-_7245152 | 0.17 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr4_+_74606223 | 0.17 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr12_+_28605426 | 0.16 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_10007448 | 0.16 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr3_-_139258521 | 0.16 |
ENST00000483943.2
ENST00000232219.2 ENST00000492918.1 |
RBP1
|
retinol binding protein 1, cellular |
| chr2_-_118943930 | 0.16 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr19_-_51143075 | 0.16 |
ENST00000600079.1
ENST00000593901.1 |
SYT3
|
synaptotagmin III |
| chr6_+_131894284 | 0.15 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr14_+_39583427 | 0.15 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr17_-_54893250 | 0.15 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chrX_-_77150911 | 0.15 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr14_+_65878650 | 0.14 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr12_+_19358192 | 0.14 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr2_+_210518057 | 0.14 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr11_-_104972158 | 0.14 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr9_-_75488984 | 0.14 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr6_+_32812568 | 0.14 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr22_-_19466643 | 0.14 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr22_+_19118321 | 0.13 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr2_-_70417827 | 0.13 |
ENST00000457952.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr7_+_120629653 | 0.13 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr4_+_186990298 | 0.13 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr19_+_50148087 | 0.13 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chrX_-_109590174 | 0.13 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr15_-_49913126 | 0.13 |
ENST00000561064.1
ENST00000299338.6 |
FAM227B
|
family with sequence similarity 227, member B |
| chr8_+_52730143 | 0.13 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr7_+_6713376 | 0.12 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr5_+_135496675 | 0.12 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr2_-_55647057 | 0.12 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr14_+_65878565 | 0.12 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr14_+_62164340 | 0.12 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr10_+_17794251 | 0.11 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr4_-_185395191 | 0.11 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr7_-_92146729 | 0.11 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr2_+_143635222 | 0.11 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr2_-_158182105 | 0.10 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr5_+_127039075 | 0.10 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr6_+_1312675 | 0.10 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chr3_+_107602030 | 0.10 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr1_+_212475148 | 0.10 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr1_+_16083098 | 0.10 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr5_+_140588269 | 0.10 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr13_-_44735393 | 0.10 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr8_+_119294456 | 0.09 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr2_-_158182322 | 0.09 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr20_+_5987890 | 0.09 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr4_+_69962212 | 0.09 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_+_202385953 | 0.09 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_-_76944621 | 0.09 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr16_+_53483983 | 0.09 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr11_+_12115543 | 0.09 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr2_-_55646957 | 0.09 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr8_-_90993869 | 0.09 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr18_+_21452804 | 0.08 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr1_+_219347203 | 0.08 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr1_+_149239529 | 0.08 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr20_+_9146969 | 0.08 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr11_+_123986069 | 0.08 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr5_-_147211226 | 0.08 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr5_-_65018834 | 0.08 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr13_+_76413852 | 0.08 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr6_-_31632962 | 0.08 |
ENST00000456540.1
ENST00000445768.1 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr6_+_24775153 | 0.08 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr1_+_45140360 | 0.08 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr18_-_57027194 | 0.08 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chrY_+_14958970 | 0.08 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr11_+_69924639 | 0.08 |
ENST00000538023.1
ENST00000398543.2 |
ANO1
|
anoctamin 1, calcium activated chloride channel |
| chr8_+_79503458 | 0.07 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr4_+_37455536 | 0.07 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chrX_+_36246735 | 0.07 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr6_+_130339710 | 0.07 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr7_-_122339162 | 0.07 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr6_-_133055815 | 0.07 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr17_+_9479944 | 0.07 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr6_+_131958436 | 0.07 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr12_-_91546926 | 0.07 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_-_213020991 | 0.07 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr2_+_145780739 | 0.07 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_-_46639436 | 0.07 |
ENST00000532281.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr4_-_168155730 | 0.07 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_+_39640787 | 0.07 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr15_+_58430368 | 0.07 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr10_-_115614127 | 0.07 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr5_-_111093167 | 0.07 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr2_+_114647504 | 0.06 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr6_-_49712123 | 0.06 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr17_+_61086917 | 0.06 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_-_131321863 | 0.06 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr3_+_140981456 | 0.06 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr2_+_143886877 | 0.06 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr18_-_74728998 | 0.06 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr14_+_62462541 | 0.06 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr4_+_169575875 | 0.06 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr13_-_33924755 | 0.06 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr21_-_39705323 | 0.06 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr22_+_50609150 | 0.06 |
ENST00000159647.5
ENST00000395842.2 |
PANX2
|
pannexin 2 |
| chr21_+_35736302 | 0.06 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr1_+_198608146 | 0.06 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr6_-_28220002 | 0.06 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr3_-_107777208 | 0.06 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr12_-_9268707 | 0.05 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr1_+_104615595 | 0.05 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chrX_+_36254051 | 0.05 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr13_+_51483814 | 0.05 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr19_-_5903714 | 0.05 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr15_+_58430567 | 0.05 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr11_-_102401469 | 0.05 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chrX_-_77150985 | 0.05 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr5_-_96518907 | 0.05 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr11_+_107461804 | 0.05 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr7_-_6866401 | 0.05 |
ENST00000316731.8
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr2_+_210444748 | 0.05 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr10_-_105845674 | 0.05 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chrX_-_72434628 | 0.05 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr11_-_128457446 | 0.05 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr1_-_244006528 | 0.05 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr12_+_19358228 | 0.05 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_-_157221380 | 0.05 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr8_+_86089460 | 0.05 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr17_-_71223839 | 0.05 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr11_-_8986474 | 0.05 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
| chr2_+_234668894 | 0.04 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr16_+_22518495 | 0.04 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chrX_+_71401570 | 0.04 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr5_-_138775177 | 0.04 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr6_+_149539767 | 0.04 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr14_+_92789498 | 0.04 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr2_+_109237717 | 0.04 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_-_48937838 | 0.04 |
ENST00000371847.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr3_+_111393501 | 0.04 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr12_-_10607084 | 0.04 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr1_+_81771806 | 0.04 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr5_+_112043186 | 0.04 |
ENST00000509732.1
ENST00000457016.1 ENST00000507379.1 |
APC
|
adenomatous polyposis coli |
| chr6_-_88411911 | 0.04 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr17_+_75181292 | 0.04 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr1_-_48937821 | 0.04 |
ENST00000396199.3
|
SPATA6
|
spermatogenesis associated 6 |
| chr8_-_102181718 | 0.04 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr4_-_168155577 | 0.04 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_+_4087664 | 0.04 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr4_+_74576019 | 0.04 |
ENST00000436089.1
|
AC112518.3
|
AC112518.3 |
| chr15_-_75748115 | 0.04 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr14_-_68000442 | 0.03 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr12_-_95009837 | 0.03 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr20_-_39928756 | 0.03 |
ENST00000432768.2
|
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr1_+_67632083 | 0.03 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chrX_+_135388147 | 0.03 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr10_-_94301107 | 0.03 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr1_+_160370344 | 0.03 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_+_161993412 | 0.03 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr9_+_20927764 | 0.03 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr12_-_30887948 | 0.03 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr3_+_62936098 | 0.03 |
ENST00000475886.1
ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698
|
long intergenic non-protein coding RNA 698 |
| chr15_-_54025300 | 0.03 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chrX_+_12993336 | 0.03 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chrX_-_10645724 | 0.03 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr15_+_48483736 | 0.03 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr18_+_616711 | 0.03 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr5_+_137722255 | 0.03 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr9_-_28670283 | 0.03 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr3_+_182511266 | 0.02 |
ENST00000323116.5
ENST00000493826.1 |
ATP11B
|
ATPase, class VI, type 11B |
| chr1_+_197881592 | 0.02 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr9_+_124103625 | 0.02 |
ENST00000594963.1
|
AL161784.1
|
Uncharacterized protein |
| chr4_-_69817481 | 0.02 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr15_+_52155001 | 0.02 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr5_-_177207634 | 0.02 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr9_+_96846740 | 0.02 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr3_+_102153859 | 0.02 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr10_-_99030395 | 0.02 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr8_-_10512569 | 0.02 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr18_-_24443151 | 0.02 |
ENST00000440832.3
|
AQP4
|
aquaporin 4 |
| chr12_-_15815626 | 0.02 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr15_+_57998923 | 0.02 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr22_-_32651326 | 0.02 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr7_+_93535817 | 0.02 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr12_+_96196875 | 0.02 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chrX_+_36053908 | 0.02 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr1_-_146082633 | 0.02 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr7_+_77469439 | 0.02 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0015254 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |