Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
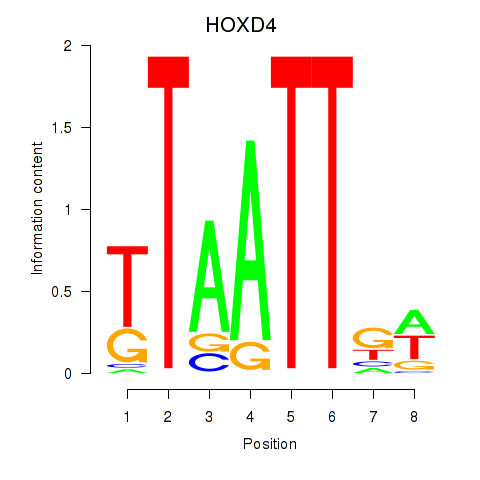
Results for HOXD4
Z-value: 0.67
Transcription factors associated with HOXD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD4
|
ENSG00000170166.5 | homeobox D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD4 | hg19_v2_chr2_+_177015950_177015950 | -0.13 | 8.7e-01 | Click! |
Activity profile of HOXD4 motif
Sorted Z-values of HOXD4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_89442940 | 1.23 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr2_+_179318295 | 1.05 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr17_-_64225508 | 0.95 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_155484103 | 0.93 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484155 | 0.87 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr5_+_31193847 | 0.74 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr3_+_142342228 | 0.60 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr3_-_195310802 | 0.57 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr13_-_54706954 | 0.42 |
ENST00000606706.1
ENST00000607494.1 ENST00000427299.2 ENST00000423442.2 ENST00000451744.1 |
LINC00458
|
long intergenic non-protein coding RNA 458 |
| chr13_+_37581115 | 0.40 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr4_-_76944621 | 0.37 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr7_+_77428066 | 0.37 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr9_+_97562440 | 0.33 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr7_+_16793160 | 0.33 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr8_-_102803163 | 0.32 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr2_+_33701684 | 0.32 |
ENST00000442390.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_+_56590002 | 0.32 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr5_+_68860949 | 0.32 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr5_+_111755280 | 0.32 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr5_-_111754948 | 0.30 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr6_+_130339710 | 0.30 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr15_-_55700457 | 0.29 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr13_+_110958124 | 0.27 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr16_-_3350614 | 0.27 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr5_+_95066823 | 0.26 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr15_+_36887069 | 0.25 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr2_+_179317994 | 0.24 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr4_-_130014729 | 0.23 |
ENST00000281142.5
ENST00000434680.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr1_+_186798073 | 0.23 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr7_-_104909435 | 0.23 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr4_-_105416039 | 0.23 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr2_+_190541153 | 0.22 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr8_+_42873548 | 0.22 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr7_+_77428149 | 0.22 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr12_-_39734783 | 0.22 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr2_-_160473114 | 0.22 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr8_-_27941380 | 0.21 |
ENST00000413272.2
ENST00000341513.6 |
NUGGC
|
nuclear GTPase, germinal center associated |
| chr17_+_9479944 | 0.21 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr4_+_169753156 | 0.21 |
ENST00000393726.3
ENST00000507735.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr9_+_130026756 | 0.21 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr18_-_21891460 | 0.20 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chrX_+_77166172 | 0.20 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr3_-_149095652 | 0.20 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr15_-_55700522 | 0.19 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr7_+_135611542 | 0.19 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr15_-_54025300 | 0.19 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr12_-_94673956 | 0.19 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr4_+_169418195 | 0.18 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr20_-_45980621 | 0.18 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_-_101360331 | 0.18 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr2_-_225434538 | 0.17 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr8_-_42396185 | 0.17 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr15_+_93443419 | 0.17 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr3_-_11623804 | 0.17 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_-_101360374 | 0.17 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr2_+_158114051 | 0.16 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr13_+_20207782 | 0.16 |
ENST00000414242.2
ENST00000361479.5 |
MPHOSPH8
|
M-phase phosphoprotein 8 |
| chr6_-_36515177 | 0.16 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr5_-_54988448 | 0.15 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr16_-_15736953 | 0.15 |
ENST00000548025.1
ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430
|
KIAA0430 |
| chr2_+_170440844 | 0.15 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr6_-_136847099 | 0.15 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr16_+_87636474 | 0.14 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr2_+_187371440 | 0.14 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr4_+_124571409 | 0.14 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr14_+_61449076 | 0.13 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr4_-_152149033 | 0.13 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr21_-_40984704 | 0.13 |
ENST00000329618.6
|
C21orf88
|
chromosome 21 open reading frame 88 |
| chr6_-_32157947 | 0.12 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr1_-_87379785 | 0.12 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr1_-_87380002 | 0.12 |
ENST00000331835.5
|
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr12_-_112279694 | 0.12 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr11_+_12766583 | 0.12 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr13_+_73629107 | 0.12 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr3_+_97483572 | 0.12 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr17_+_59489112 | 0.12 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr17_-_29624343 | 0.12 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr9_-_128246769 | 0.12 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr10_-_14372870 | 0.11 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr20_+_18447771 | 0.11 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr10_+_86184676 | 0.11 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr22_-_29107919 | 0.11 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr14_+_99947715 | 0.11 |
ENST00000389879.5
ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK
|
cyclin K |
| chr7_-_77427676 | 0.11 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr15_-_65809991 | 0.10 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr10_+_95848824 | 0.10 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr11_-_31531121 | 0.09 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_16109519 | 0.09 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr4_-_130014705 | 0.09 |
ENST00000503401.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chr1_+_203444887 | 0.09 |
ENST00000343110.2
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein |
| chrX_+_37850026 | 0.09 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr4_-_130014532 | 0.08 |
ENST00000506368.1
ENST00000439369.2 ENST00000503215.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr14_+_90863327 | 0.08 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr2_-_99279928 | 0.08 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr3_-_4508925 | 0.08 |
ENST00000534863.1
ENST00000383843.5 ENST00000458465.2 ENST00000405420.2 ENST00000272902.5 |
SUMF1
|
sulfatase modifying factor 1 |
| chrX_+_70752917 | 0.08 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr19_+_30433110 | 0.08 |
ENST00000542441.2
ENST00000392271.1 |
URI1
|
URI1, prefoldin-like chaperone |
| chr6_-_53013620 | 0.08 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr8_-_42698433 | 0.08 |
ENST00000345117.2
ENST00000254250.3 |
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr5_-_179499108 | 0.08 |
ENST00000521389.1
|
RNF130
|
ring finger protein 130 |
| chr18_+_32556892 | 0.08 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr16_+_84801852 | 0.08 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr14_+_62164340 | 0.08 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr19_+_50016610 | 0.08 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_102714534 | 0.07 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr8_+_107738343 | 0.07 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr3_-_178103144 | 0.07 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr4_-_130014609 | 0.07 |
ENST00000511426.1
|
SCLT1
|
sodium channel and clathrin linker 1 |
| chrX_+_37865804 | 0.06 |
ENST00000297875.2
ENST00000357972.5 |
SYTL5
|
synaptotagmin-like 5 |
| chr5_+_141346385 | 0.06 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr5_+_59783941 | 0.06 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_+_201617450 | 0.06 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr3_+_69812877 | 0.06 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr3_+_158787041 | 0.06 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr17_+_47448102 | 0.06 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr6_+_142468361 | 0.05 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr1_-_151762900 | 0.05 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr1_-_231560790 | 0.05 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr2_-_159237472 | 0.05 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr2_-_27579842 | 0.05 |
ENST00000423998.1
ENST00000264720.3 |
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr2_-_31361543 | 0.04 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr10_+_13652047 | 0.04 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr5_-_179499086 | 0.04 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr7_+_112063192 | 0.04 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr21_+_35736302 | 0.04 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr16_+_24549014 | 0.04 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr5_+_67586465 | 0.04 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_39979576 | 0.04 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr5_+_66300446 | 0.04 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_+_24151553 | 0.04 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr16_-_29910853 | 0.04 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr12_-_86650077 | 0.03 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_-_61311485 | 0.03 |
ENST00000436264.1
ENST00000356424.6 ENST00000341074.5 |
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr13_+_51913819 | 0.03 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr4_-_143227088 | 0.03 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_-_22233442 | 0.03 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr20_+_5931497 | 0.03 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr1_+_84630645 | 0.03 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_+_21168630 | 0.03 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_-_208030295 | 0.03 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_170115142 | 0.03 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr20_+_32150140 | 0.02 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr4_-_46996424 | 0.02 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr1_+_144146808 | 0.02 |
ENST00000369190.5
ENST00000412624.2 ENST00000369365.3 |
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr10_-_61899124 | 0.02 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr6_-_44281043 | 0.02 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr17_+_39382900 | 0.02 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr2_+_234590556 | 0.02 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr11_-_36310958 | 0.02 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr4_-_153601136 | 0.02 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr1_+_87380299 | 0.02 |
ENST00000370551.4
ENST00000370550.5 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr5_-_150727111 | 0.02 |
ENST00000335244.4
ENST00000521967.1 |
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2 |
| chr22_+_23248512 | 0.02 |
ENST00000390325.2
|
IGLC3
|
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr15_-_64386120 | 0.02 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr18_-_74839891 | 0.02 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr2_+_224822121 | 0.02 |
ENST00000258383.3
|
MRPL44
|
mitochondrial ribosomal protein L44 |
| chr5_-_110848189 | 0.01 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr18_+_22040593 | 0.01 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr5_+_66300464 | 0.01 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_112041253 | 0.01 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr6_-_76072719 | 0.01 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr5_+_179921344 | 0.01 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr3_-_149293990 | 0.01 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chrX_+_139791917 | 0.01 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr14_-_24911868 | 0.01 |
ENST00000554698.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr11_-_77790865 | 0.01 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr5_+_157158205 | 0.00 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr8_-_124279627 | 0.00 |
ENST00000357082.4
|
ZHX1-C8ORF76
|
ZHX1-C8ORF76 readthrough |
| chr9_+_273038 | 0.00 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr2_+_12246664 | 0.00 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr2_-_183291741 | 0.00 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 1.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 1.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.3 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |