Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
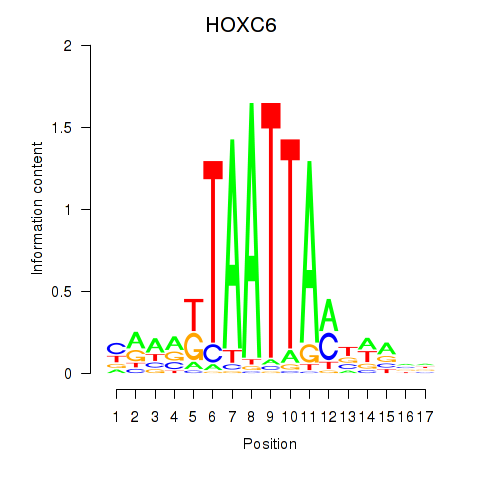
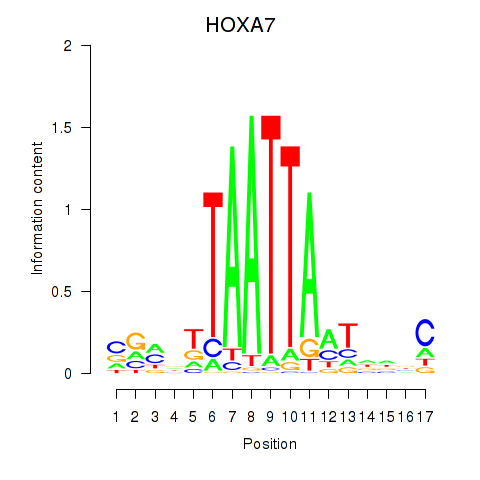
Results for HOXC6_HOXA7
Z-value: 1.12
Transcription factors associated with HOXC6_HOXA7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC6
|
ENSG00000197757.7 | homeobox C6 |
|
HOXA7
|
ENSG00000122592.6 | homeobox A7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC6 | hg19_v2_chr12_+_54422142_54422239 | 0.90 | 1.0e-01 | Click! |
| HOXA7 | hg19_v2_chr7_-_27196267_27196311 | 0.40 | 6.0e-01 | Click! |
Activity profile of HOXC6_HOXA7 motif
Sorted Z-values of HOXC6_HOXA7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_12059091 | 1.81 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_+_155484103 | 1.72 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484155 | 1.62 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr19_-_36304201 | 1.35 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr3_+_149191723 | 1.26 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr14_-_94789663 | 1.23 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr17_-_64225508 | 1.12 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr10_-_101825151 | 1.10 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr6_-_109702885 | 0.97 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr17_+_1674982 | 0.85 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_-_91573132 | 0.68 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr15_+_69857515 | 0.67 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr15_-_55657428 | 0.62 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr8_-_124741451 | 0.61 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr17_-_4938712 | 0.60 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr8_+_27238147 | 0.59 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr10_-_27529486 | 0.56 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_+_108745711 | 0.52 |
ENST00000394684.4
|
SGMS2
|
sphingomyelin synthase 2 |
| chr6_+_26365443 | 0.50 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr3_-_149093499 | 0.50 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chrX_+_43515467 | 0.48 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr19_+_49199209 | 0.47 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr2_+_30670077 | 0.47 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr19_+_50016610 | 0.46 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_+_57233087 | 0.45 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr11_-_111649015 | 0.45 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr7_-_111032971 | 0.44 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr14_-_90085458 | 0.41 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr5_+_95066823 | 0.41 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr12_-_112279694 | 0.40 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr11_+_86748863 | 0.39 |
ENST00000340353.7
|
TMEM135
|
transmembrane protein 135 |
| chr6_-_33160231 | 0.39 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr17_+_48823975 | 0.37 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr9_+_125133315 | 0.35 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr6_+_26365387 | 0.34 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr9_-_5339873 | 0.33 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr16_-_28937027 | 0.33 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr12_-_89920030 | 0.33 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr13_+_36050881 | 0.33 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr4_+_169418195 | 0.33 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr14_+_50234309 | 0.32 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr4_+_113568207 | 0.32 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr6_+_130339710 | 0.30 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr12_-_89919765 | 0.30 |
ENST00000541909.1
ENST00000313546.3 |
POC1B
|
POC1 centriolar protein B |
| chr2_-_134326009 | 0.29 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr10_+_99205959 | 0.29 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr6_-_47445214 | 0.28 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr21_-_43346790 | 0.28 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr2_+_132479948 | 0.27 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chrX_-_106362013 | 0.27 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr19_-_51920952 | 0.27 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr11_-_8857248 | 0.26 |
ENST00000534248.1
ENST00000530959.1 ENST00000531578.1 ENST00000533225.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr11_+_66276550 | 0.26 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr7_-_55620433 | 0.25 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr15_+_35270552 | 0.25 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr16_-_28634874 | 0.24 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_+_95128748 | 0.24 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_+_112679301 | 0.24 |
ENST00000265277.5
ENST00000369452.4 |
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans) |
| chr6_-_36515177 | 0.23 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr17_-_46688334 | 0.23 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr15_-_75748143 | 0.22 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr10_+_99205894 | 0.22 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr6_+_28092338 | 0.22 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr5_+_162930114 | 0.22 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr1_+_221051699 | 0.22 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr4_-_39979576 | 0.22 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr6_+_96969672 | 0.21 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr12_+_79371565 | 0.21 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr19_+_50016411 | 0.21 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_+_117544366 | 0.21 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr7_+_148936732 | 0.20 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr1_-_242612779 | 0.20 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr15_+_96904487 | 0.20 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr4_-_185275104 | 0.19 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr14_-_23426231 | 0.19 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr17_-_76220740 | 0.18 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr6_+_26440700 | 0.18 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr14_-_23426270 | 0.17 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr17_+_35851570 | 0.17 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr18_+_32558208 | 0.17 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_-_101989315 | 0.16 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr20_+_15177480 | 0.16 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr1_+_202317855 | 0.16 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_-_100009856 | 0.15 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr1_+_84609944 | 0.15 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_+_136649311 | 0.15 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr6_-_38670897 | 0.15 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr4_-_152149033 | 0.15 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr14_-_23426337 | 0.14 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr9_-_215744 | 0.14 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr7_-_32338917 | 0.14 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr7_+_134671234 | 0.14 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr3_+_152552685 | 0.14 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr1_+_66820058 | 0.14 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr14_-_23426322 | 0.14 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chrX_+_9431324 | 0.13 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr5_+_140625147 | 0.13 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr7_-_130080977 | 0.13 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr4_-_170533723 | 0.13 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr9_-_27005686 | 0.13 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chrX_-_16887963 | 0.13 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr6_-_34639733 | 0.12 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr5_-_89770582 | 0.12 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr13_-_74993252 | 0.11 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr11_+_126173647 | 0.10 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr8_-_82373809 | 0.10 |
ENST00000379071.2
|
FABP9
|
fatty acid binding protein 9, testis |
| chr3_-_141719195 | 0.09 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_-_94673956 | 0.09 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr2_-_176046391 | 0.09 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr7_+_134528635 | 0.09 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr6_+_30029008 | 0.09 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr6_+_149721495 | 0.09 |
ENST00000326669.4
|
SUMO4
|
small ubiquitin-like modifier 4 |
| chr16_+_53469525 | 0.08 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr6_+_114178512 | 0.08 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr1_+_212965170 | 0.08 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr21_-_35796241 | 0.08 |
ENST00000450895.1
|
AP000322.53
|
AP000322.53 |
| chr11_-_559377 | 0.07 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr5_+_54455946 | 0.07 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr4_+_71019903 | 0.07 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr16_+_28565230 | 0.07 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr14_+_31046959 | 0.06 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr8_-_133772870 | 0.06 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chrM_+_10053 | 0.06 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr12_+_56435637 | 0.06 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chrM_+_10464 | 0.06 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr2_-_74618907 | 0.06 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr17_-_39623681 | 0.06 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr8_+_77593474 | 0.05 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr2_-_128284020 | 0.05 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr12_-_86650077 | 0.05 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_-_142888573 | 0.04 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr1_+_62439037 | 0.04 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr13_-_30881134 | 0.04 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr12_+_131438443 | 0.04 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr9_-_95166841 | 0.04 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr10_-_112678692 | 0.04 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr1_+_117963209 | 0.04 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr7_+_141811539 | 0.04 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr3_+_157154578 | 0.04 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr18_+_32558380 | 0.04 |
ENST00000588349.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_202317815 | 0.04 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_112678904 | 0.04 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr9_-_34329198 | 0.04 |
ENST00000379166.2
ENST00000345050.2 |
KIF24
|
kinesin family member 24 |
| chr2_+_234637754 | 0.04 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_+_138340067 | 0.04 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_77790865 | 0.03 |
ENST00000534029.1
ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2
NDUFC2-KCTD14
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr11_+_112041253 | 0.03 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr16_+_15489629 | 0.03 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr2_+_234621551 | 0.03 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr12_+_60083118 | 0.03 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_-_196911002 | 0.03 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr14_+_56584414 | 0.03 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr12_-_50643664 | 0.03 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr11_-_31391276 | 0.03 |
ENST00000452803.1
|
DCDC1
|
doublecortin domain containing 1 |
| chr13_+_31309645 | 0.02 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr12_+_26348429 | 0.02 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr10_-_74283694 | 0.02 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr6_+_4087664 | 0.02 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chrX_-_14047996 | 0.01 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr4_-_159956333 | 0.01 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr10_-_112678976 | 0.01 |
ENST00000448814.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr11_-_49230184 | 0.01 |
ENST00000340334.7
ENST00000256999.2 |
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chr13_+_20268547 | 0.01 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr5_+_66300446 | 0.01 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_22862448 | 0.01 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr5_-_86534822 | 0.01 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr4_+_169418255 | 0.01 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_-_87281196 | 0.01 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr20_-_50384864 | 0.00 |
ENST00000311637.5
ENST00000402822.1 |
ATP9A
|
ATPase, class II, type 9A |
| chr18_-_74839891 | 0.00 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr4_-_87281224 | 0.00 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC6_HOXA7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 3.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.7 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.2 | 1.4 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.8 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.6 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 1.2 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:1902739 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 1.8 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 3.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |