Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
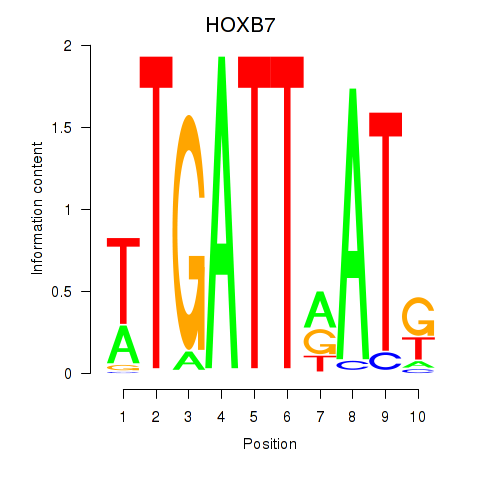
Results for HOXB7
Z-value: 1.06
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | -0.95 | 4.9e-02 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102668879 | 1.17 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr2_+_169926047 | 0.84 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr2_+_176972000 | 0.84 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr11_+_57365150 | 0.68 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr7_-_105926058 | 0.68 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr3_-_185270342 | 0.67 |
ENST00000424591.2
|
LIPH
|
lipase, member H |
| chr17_+_7758374 | 0.65 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr16_+_15596123 | 0.64 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr17_+_58018269 | 0.59 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr14_+_101299520 | 0.57 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_-_74800799 | 0.57 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chrX_-_1331527 | 0.57 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr14_+_55034599 | 0.54 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr18_+_21464737 | 0.54 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr4_-_120243545 | 0.54 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr9_-_32526299 | 0.52 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chrX_+_1734051 | 0.52 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr12_+_43086018 | 0.50 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr4_-_141075330 | 0.49 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr14_+_101295638 | 0.47 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_+_5646213 | 0.46 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr15_-_60683326 | 0.45 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr10_+_13142075 | 0.42 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr3_-_4927447 | 0.42 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr13_-_103719196 | 0.41 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr7_-_33080506 | 0.41 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr22_-_30642728 | 0.39 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr1_+_104615595 | 0.39 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr14_-_101295407 | 0.39 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr4_+_169013666 | 0.38 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr22_-_30642782 | 0.37 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr14_+_35591858 | 0.37 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chr10_+_13141585 | 0.37 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr22_+_46449674 | 0.36 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr12_+_40787194 | 0.36 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr10_+_13142225 | 0.35 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr14_-_65769392 | 0.35 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr4_+_26323764 | 0.35 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_51952418 | 0.34 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr15_+_67418047 | 0.34 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr3_+_173116225 | 0.34 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr4_-_44653636 | 0.33 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr6_-_51952367 | 0.32 |
ENST00000340994.4
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr17_+_76494911 | 0.32 |
ENST00000598378.1
|
DNAH17-AS1
|
DNAH17 antisense RNA 1 |
| chr10_+_13141441 | 0.32 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr2_-_70780770 | 0.32 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr1_-_92952433 | 0.32 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr2_-_99917639 | 0.32 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr11_-_111649074 | 0.31 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr10_+_47894572 | 0.31 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chrX_-_77225135 | 0.31 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr4_-_72649763 | 0.30 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr8_-_67874805 | 0.30 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr17_+_67498295 | 0.30 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_+_107650219 | 0.30 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr11_-_57194919 | 0.29 |
ENST00000532795.1
|
SLC43A3
|
solute carrier family 43, member 3 |
| chr3_-_108836945 | 0.29 |
ENST00000483760.1
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr1_+_101702417 | 0.29 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr12_-_91546926 | 0.29 |
ENST00000550758.1
|
DCN
|
decorin |
| chr9_-_73029540 | 0.28 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr5_+_67485704 | 0.28 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr4_-_77328458 | 0.28 |
ENST00000388914.3
ENST00000434846.2 |
CCDC158
|
coiled-coil domain containing 158 |
| chr7_+_120629653 | 0.28 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_-_137539651 | 0.28 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr9_-_75488984 | 0.28 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr11_-_70507901 | 0.28 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr9_+_74729511 | 0.28 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr4_+_74301880 | 0.27 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr1_-_224624730 | 0.27 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr16_+_67880574 | 0.27 |
ENST00000219169.4
|
NUTF2
|
nuclear transport factor 2 |
| chr11_+_44117219 | 0.27 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr10_+_5406935 | 0.27 |
ENST00000380433.3
|
UCN3
|
urocortin 3 |
| chr6_+_54172653 | 0.27 |
ENST00000370869.3
|
TINAG
|
tubulointerstitial nephritis antigen |
| chr4_+_26324474 | 0.27 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_+_82660635 | 0.26 |
ENST00000567445.1
ENST00000446376.2 |
CDH13
|
cadherin 13 |
| chr21_+_44313375 | 0.26 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr8_-_66750978 | 0.26 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
| chr11_-_57194948 | 0.26 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr3_+_191046810 | 0.26 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr1_-_246357029 | 0.26 |
ENST00000391836.2
|
SMYD3
|
SET and MYND domain containing 3 |
| chr16_+_20911174 | 0.26 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr19_-_56709162 | 0.26 |
ENST00000589938.1
ENST00000587032.2 ENST00000586855.2 |
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr12_+_28605426 | 0.26 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr18_-_51750948 | 0.26 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr7_+_135777671 | 0.25 |
ENST00000445293.2
ENST00000435996.1 |
AC009784.3
|
AC009784.3 |
| chr10_+_47894023 | 0.25 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr8_-_116681123 | 0.25 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_+_45464658 | 0.25 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chr1_+_152178320 | 0.25 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr13_-_41240717 | 0.25 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr12_+_59989918 | 0.25 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_+_140227357 | 0.24 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr19_-_30199516 | 0.24 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr2_-_154335300 | 0.24 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr14_+_77425972 | 0.23 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr10_+_124320156 | 0.23 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chrX_+_13707235 | 0.23 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr10_+_69869237 | 0.23 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr15_-_42343388 | 0.23 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr16_-_68034470 | 0.23 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr10_+_71561630 | 0.23 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr1_+_112016414 | 0.23 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr12_-_22063787 | 0.22 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr6_-_24720226 | 0.22 |
ENST00000378102.3
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr10_+_24755416 | 0.22 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr14_-_24584138 | 0.22 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr16_+_22501658 | 0.22 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chrX_-_110655306 | 0.22 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr6_-_66417107 | 0.22 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr21_-_33984456 | 0.22 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr1_+_205473720 | 0.22 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr16_-_66583994 | 0.22 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr1_-_91487806 | 0.21 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr12_+_113229737 | 0.21 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr3_-_169587621 | 0.21 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr1_+_244227632 | 0.21 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chrX_-_106243294 | 0.21 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr12_-_11339543 | 0.21 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr17_-_7218631 | 0.21 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr7_+_116654958 | 0.21 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_112929850 | 0.21 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr7_-_7782204 | 0.21 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr19_+_35417939 | 0.21 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr7_-_3214287 | 0.21 |
ENST00000404626.3
|
AC091801.1
|
LOC392621; Uncharacterized protein |
| chr17_+_44803922 | 0.20 |
ENST00000465370.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr19_+_54369608 | 0.20 |
ENST00000336967.3
|
MYADM
|
myeloid-associated differentiation marker |
| chr14_-_54425475 | 0.20 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr8_-_29208183 | 0.20 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chrX_+_1387693 | 0.20 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr12_+_78359999 | 0.20 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr5_-_173217931 | 0.20 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr15_+_64680003 | 0.20 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr11_-_63376013 | 0.20 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr4_+_69962212 | 0.20 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr17_-_46507567 | 0.20 |
ENST00000584924.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr2_-_200322723 | 0.19 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr12_+_59989791 | 0.19 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_61196858 | 0.19 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_+_120303717 | 0.19 |
ENST00000594371.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr10_-_51958906 | 0.19 |
ENST00000489640.1
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr2_+_27886330 | 0.19 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr5_+_131409476 | 0.19 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr12_+_123944070 | 0.18 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr11_+_35222629 | 0.18 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr2_+_39117010 | 0.18 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr1_+_66999799 | 0.18 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr7_+_2636522 | 0.18 |
ENST00000423196.1
|
IQCE
|
IQ motif containing E |
| chr6_+_34204642 | 0.18 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_+_234826016 | 0.18 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr12_+_113229543 | 0.18 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr16_-_63651939 | 0.18 |
ENST00000563855.1
ENST00000568932.1 ENST00000564290.1 |
RP11-368L12.1
|
RP11-368L12.1 |
| chr1_+_224370873 | 0.18 |
ENST00000323699.4
ENST00000391877.3 |
DEGS1
|
delta(4)-desaturase, sphingolipid 1 |
| chr21_+_43619796 | 0.18 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr9_+_105757590 | 0.17 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_-_145878954 | 0.17 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr3_-_108836977 | 0.17 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr19_+_41698927 | 0.17 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr1_-_178840157 | 0.17 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr17_+_60447579 | 0.17 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr8_+_12803176 | 0.17 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr3_+_145782358 | 0.17 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr20_+_10199566 | 0.17 |
ENST00000430336.1
|
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr12_+_101988627 | 0.17 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr18_+_3252206 | 0.17 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr7_-_120498357 | 0.17 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr12_+_101988774 | 0.17 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr8_+_28748765 | 0.17 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr20_-_1309809 | 0.17 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr17_-_46703826 | 0.17 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr11_-_107729887 | 0.17 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr3_-_183145873 | 0.16 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr16_-_18462221 | 0.16 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr4_-_111558135 | 0.16 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chrX_-_92928557 | 0.16 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chrX_+_36053908 | 0.16 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr8_-_116681686 | 0.16 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr13_+_108921977 | 0.16 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr18_+_22040620 | 0.16 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr4_+_158493642 | 0.16 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr5_+_125758813 | 0.16 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_-_106243451 | 0.16 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr19_+_20959098 | 0.15 |
ENST00000360204.5
ENST00000594534.1 |
ZNF66
|
zinc finger protein 66 |
| chr17_-_69198295 | 0.15 |
ENST00000569074.1
|
CASC17
|
cancer susceptibility candidate 17 (non-protein coding) |
| chr11_-_107729287 | 0.15 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr14_-_23624511 | 0.15 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr16_-_88729473 | 0.15 |
ENST00000301012.3
ENST00000569177.1 |
MVD
|
mevalonate (diphospho) decarboxylase |
| chr5_-_58295712 | 0.15 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_90025011 | 0.15 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr5_+_125758865 | 0.15 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr16_+_82660560 | 0.15 |
ENST00000268613.10
ENST00000565636.1 ENST00000431540.3 ENST00000428848.3 |
CDH13
|
cadherin 13 |
| chr2_+_157330081 | 0.15 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr14_-_68159152 | 0.15 |
ENST00000554035.1
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
| chr12_+_113229452 | 0.15 |
ENST00000389385.4
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr3_+_159943362 | 0.15 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr8_-_67090825 | 0.14 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr12_+_21168630 | 0.14 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr8_+_104892639 | 0.14 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_219110149 | 0.14 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr17_-_39203519 | 0.14 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr5_-_33297946 | 0.14 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.5 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 0.8 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.5 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.4 | GO:0044115 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.3 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.5 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.6 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.8 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.2 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.4 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.2 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.2 | GO:2000004 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.7 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.2 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) tolerance induction dependent upon immune response(GO:0002461) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.3 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.2 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.1 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0097513 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 0.7 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.3 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.3 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.1 | 0.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.3 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.6 | GO:0008430 | selenium binding(GO:0008430) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0016917 | GABA receptor activity(GO:0016917) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |