Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
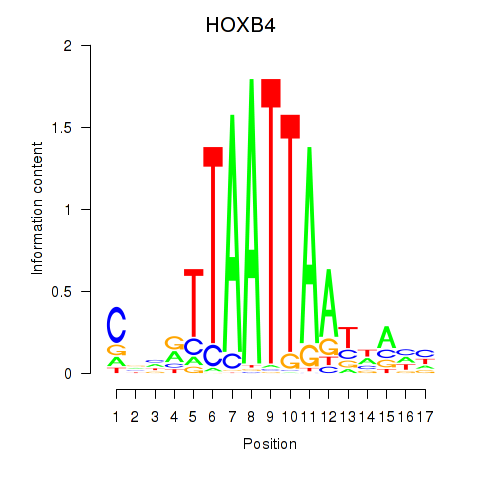
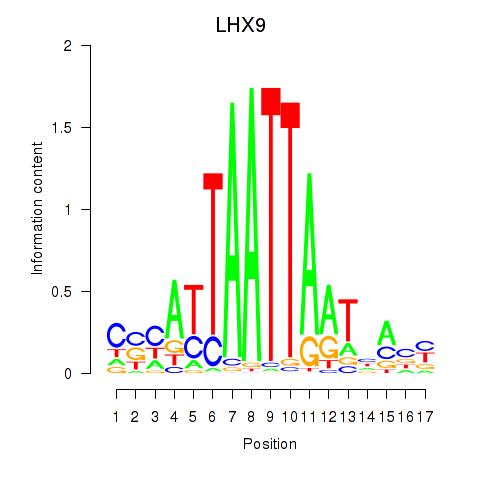
Results for HOXB4_LHX9
Z-value: 1.35
Transcription factors associated with HOXB4_LHX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB4
|
ENSG00000182742.5 | homeobox B4 |
|
LHX9
|
ENSG00000143355.11 | LIM homeobox 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX9 | hg19_v2_chr1_+_197881592_197881635 | 0.91 | 8.9e-02 | Click! |
| HOXB4 | hg19_v2_chr17_-_46657473_46657473 | -0.60 | 4.0e-01 | Click! |
Activity profile of HOXB4_LHX9 motif
Sorted Z-values of HOXB4_LHX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_5710919 | 4.01 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr19_+_54466179 | 2.80 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr11_-_327537 | 2.27 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr14_-_67878917 | 1.78 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr19_-_3557570 | 1.65 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr20_-_56265680 | 1.14 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr5_-_126409159 | 1.03 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chrX_+_10126488 | 0.94 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr15_+_64680003 | 0.92 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr15_+_90735145 | 0.87 |
ENST00000559792.1
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr18_-_51750948 | 0.87 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr1_+_225600404 | 0.86 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr1_-_212965104 | 0.85 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr12_-_10022735 | 0.85 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr22_+_46476192 | 0.84 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr2_+_182850743 | 0.81 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_182850551 | 0.80 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr11_-_121986923 | 0.80 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr12_+_38710555 | 0.79 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr5_+_179135246 | 0.79 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr14_+_24641820 | 0.78 |
ENST00000560501.1
|
REC8
|
REC8 meiotic recombination protein |
| chr14_+_24641062 | 0.76 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr11_-_74800799 | 0.76 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr8_-_90769422 | 0.72 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr4_-_120243545 | 0.71 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr9_+_22646189 | 0.65 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr5_+_89770664 | 0.64 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr16_-_66583994 | 0.62 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr21_+_33671160 | 0.61 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chrX_-_77225135 | 0.55 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr18_-_47018869 | 0.54 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr4_+_71091786 | 0.53 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr1_+_151253991 | 0.53 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr17_-_19015945 | 0.53 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr11_-_128894053 | 0.51 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr1_-_190446759 | 0.50 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr17_+_56833184 | 0.50 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr6_+_26402465 | 0.49 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr10_+_90660832 | 0.49 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr12_+_34175398 | 0.48 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr11_-_107729287 | 0.48 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_-_63376013 | 0.47 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr2_+_234826016 | 0.46 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr10_+_24755416 | 0.46 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr11_-_107729887 | 0.45 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_+_116654958 | 0.45 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr6_-_32806506 | 0.44 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr7_+_129015671 | 0.44 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr13_-_81801115 | 0.44 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chrX_-_18690210 | 0.43 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr2_-_169887827 | 0.41 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr17_+_19091325 | 0.41 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr7_-_87856303 | 0.41 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr15_+_89631647 | 0.41 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr15_+_80351910 | 0.40 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr10_+_18549645 | 0.40 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr17_+_40118773 | 0.40 |
ENST00000472031.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr1_+_155278539 | 0.40 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr20_-_1447547 | 0.39 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr8_+_39972170 | 0.39 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr16_+_15489603 | 0.38 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chrX_+_1710484 | 0.37 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr1_+_155278625 | 0.37 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr11_-_115127611 | 0.37 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chrX_+_84258832 | 0.37 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr10_+_6821545 | 0.36 |
ENST00000436383.1
|
LINC00707
|
long intergenic non-protein coding RNA 707 |
| chr20_-_1447467 | 0.36 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr17_+_33914424 | 0.36 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr7_-_87856280 | 0.36 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chrX_+_36254051 | 0.36 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr8_-_90996459 | 0.36 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr18_-_47018769 | 0.35 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr2_+_202047596 | 0.35 |
ENST00000286186.6
ENST00000360132.3 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr1_-_92952433 | 0.35 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr5_+_1225470 | 0.35 |
ENST00000324642.3
ENST00000296821.4 |
SLC6A18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr12_-_25348007 | 0.35 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr8_-_41166953 | 0.35 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr7_+_133812052 | 0.34 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr15_+_67418047 | 0.34 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_-_150020750 | 0.34 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr5_+_40841276 | 0.34 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr14_+_35591928 | 0.34 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chrX_-_110655306 | 0.33 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr4_-_119759795 | 0.33 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr5_+_89770696 | 0.33 |
ENST00000504930.1
ENST00000514483.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr3_-_141747950 | 0.32 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_26348246 | 0.32 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr7_-_150020578 | 0.32 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr10_+_99627889 | 0.32 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr7_-_25702669 | 0.32 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr11_-_124670550 | 0.31 |
ENST00000239614.4
|
MSANTD2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr1_+_160370344 | 0.31 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr16_+_82068873 | 0.31 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_+_38537960 | 0.31 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr1_+_157963063 | 0.31 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr8_-_90996837 | 0.30 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr10_-_105845674 | 0.30 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr17_+_33914460 | 0.30 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr6_+_26402517 | 0.29 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr15_+_66679155 | 0.29 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr11_-_107729504 | 0.29 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr9_-_99540328 | 0.29 |
ENST00000223428.4
ENST00000375231.1 ENST00000374641.3 |
ZNF510
|
zinc finger protein 510 |
| chr18_-_47018897 | 0.29 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr5_-_173217931 | 0.29 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr1_-_183538319 | 0.29 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr17_+_40118759 | 0.29 |
ENST00000393892.3
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr4_+_110749143 | 0.28 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr3_-_157221380 | 0.28 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr3_-_98619999 | 0.28 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr12_-_22063787 | 0.28 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr15_-_60683326 | 0.27 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr5_+_129083772 | 0.27 |
ENST00000564719.1
|
KIAA1024L
|
KIAA1024-like |
| chr12_-_112123524 | 0.26 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr6_-_135818368 | 0.26 |
ENST00000367798.2
|
AHI1
|
Abelson helper integration site 1 |
| chr6_+_42584847 | 0.26 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr15_+_48483736 | 0.26 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr3_-_87842631 | 0.26 |
ENST00000462792.1
|
RP11-451B8.1
|
RP11-451B8.1 |
| chr4_-_70518941 | 0.26 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr20_-_33735070 | 0.26 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr19_-_14064114 | 0.26 |
ENST00000585607.1
ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1
|
podocan-like 1 |
| chr19_+_4153598 | 0.25 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr11_-_111649074 | 0.25 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr16_-_29934558 | 0.25 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr14_+_103851712 | 0.25 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr12_+_93096619 | 0.24 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr18_+_21452804 | 0.24 |
ENST00000269217.6
|
LAMA3
|
laminin, alpha 3 |
| chr2_+_27886330 | 0.24 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chrX_-_15332665 | 0.24 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_+_202047843 | 0.24 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr2_+_149402989 | 0.24 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr3_-_157221357 | 0.23 |
ENST00000494677.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr14_+_55595762 | 0.23 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr11_-_104827425 | 0.23 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_142623758 | 0.23 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr7_-_149158187 | 0.22 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr12_+_78359999 | 0.22 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr15_-_72523924 | 0.22 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr14_-_104387888 | 0.22 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chr14_+_35591735 | 0.22 |
ENST00000604948.1
ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391
|
KIAA0391 |
| chr3_-_20053741 | 0.22 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr3_+_185431080 | 0.22 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr8_-_124553437 | 0.21 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr17_+_33914276 | 0.21 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_+_40841410 | 0.21 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr2_-_225811747 | 0.21 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr17_+_67498538 | 0.21 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr9_+_88556444 | 0.21 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr18_+_60206744 | 0.21 |
ENST00000586834.1
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr5_+_66300464 | 0.21 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_-_46408107 | 0.21 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr18_+_21452964 | 0.21 |
ENST00000587184.1
|
LAMA3
|
laminin, alpha 3 |
| chr19_-_36001113 | 0.21 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr1_-_57045228 | 0.20 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr1_+_152178320 | 0.20 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr6_+_31553978 | 0.20 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr4_+_158493642 | 0.20 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr20_+_18488137 | 0.20 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr8_-_116504448 | 0.20 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr4_-_103749179 | 0.19 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_64798095 | 0.19 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr1_-_47184745 | 0.19 |
ENST00000544071.1
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr1_-_168464875 | 0.19 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr6_-_33663474 | 0.19 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr11_+_74811578 | 0.19 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr2_-_158345341 | 0.19 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr12_-_25150373 | 0.19 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr9_+_125132803 | 0.18 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr10_-_105845536 | 0.18 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr16_+_3184924 | 0.18 |
ENST00000574902.1
ENST00000396878.3 |
ZNF213
|
zinc finger protein 213 |
| chr10_-_99030395 | 0.18 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr13_+_111837279 | 0.18 |
ENST00000467053.1
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_-_150473127 | 0.18 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr6_+_34204642 | 0.18 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr8_-_33424636 | 0.17 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr1_-_211307404 | 0.17 |
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr6_+_170151708 | 0.17 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr17_-_42992856 | 0.17 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr16_+_3185125 | 0.17 |
ENST00000576416.1
|
ZNF213
|
zinc finger protein 213 |
| chr2_-_88285309 | 0.17 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr14_+_35591858 | 0.17 |
ENST00000603544.1
|
KIAA0391
|
KIAA0391 |
| chr10_+_5135981 | 0.17 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr15_+_62853562 | 0.17 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr13_-_36050819 | 0.17 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr13_-_30160925 | 0.16 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr9_-_28670283 | 0.16 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_-_8000872 | 0.16 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr7_+_102553430 | 0.16 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr22_-_32767017 | 0.16 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr10_-_28571015 | 0.16 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr5_-_119669160 | 0.16 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr8_+_105235572 | 0.16 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_+_25162253 | 0.15 |
ENST00000512921.1
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr14_+_23938891 | 0.15 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chr17_+_4692230 | 0.15 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr18_-_3219847 | 0.15 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr19_-_6424783 | 0.15 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr16_+_24549014 | 0.15 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr7_+_107224364 | 0.15 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr20_+_5731083 | 0.14 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr3_-_142682178 | 0.14 |
ENST00000340634.3
|
PAQR9
|
progestin and adipoQ receptor family member IX |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB4_LHX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.2 | 0.8 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.4 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.6 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.1 | 2.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.3 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.8 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) negative regulation of heart rate(GO:0010459) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0070121 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.1 | 0.3 | GO:0090244 | negative regulation of B cell differentiation(GO:0045578) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 1.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.4 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 2.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.3 | GO:0052360 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.2 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 0.2 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 4.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.3 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.4 | GO:1904879 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 1.0 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.2 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.2 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 1.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:1901295 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:2000397 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.0 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.9 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.8 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.0 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 2.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.2 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 4.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 2.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.6 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.2 | 0.6 | GO:0070996 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.2 | 0.7 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.2 | 0.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 1.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.4 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 3.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.2 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 1.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 4.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 2.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |