Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
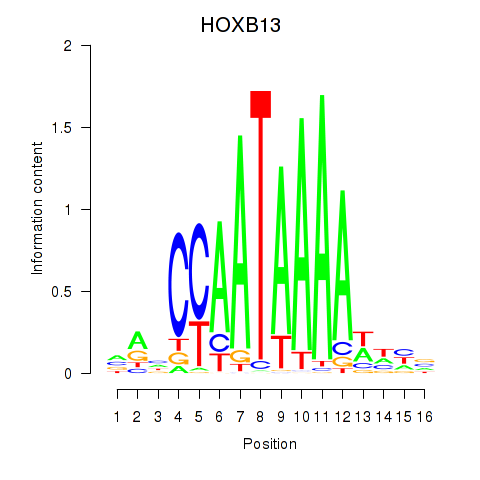
Results for HOXB13
Z-value: 0.58
Transcription factors associated with HOXB13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB13
|
ENSG00000159184.7 | homeobox B13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB13 | hg19_v2_chr17_-_46806540_46806558 | -0.92 | 8.1e-02 | Click! |
Activity profile of HOXB13 motif
Sorted Z-values of HOXB13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_126409159 | 0.68 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr8_+_86376081 | 0.64 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr1_+_23037323 | 0.47 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr5_-_145214893 | 0.47 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr15_+_66994561 | 0.36 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr8_+_119294456 | 0.33 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr3_-_114790179 | 0.33 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_155880672 | 0.30 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr19_-_5903714 | 0.30 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chrM_+_12331 | 0.29 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr18_+_68002675 | 0.25 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chrM_+_14741 | 0.24 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr7_+_35756186 | 0.24 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr17_-_39646116 | 0.23 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chrM_-_14670 | 0.21 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr7_-_50860565 | 0.20 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr17_-_73663245 | 0.19 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr1_-_36863481 | 0.19 |
ENST00000315732.2
|
LSM10
|
LSM10, U7 small nuclear RNA associated |
| chr4_+_169575875 | 0.18 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_145214848 | 0.18 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr18_+_3252206 | 0.18 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr18_-_52989525 | 0.18 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr18_+_3252265 | 0.17 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr4_+_110749143 | 0.16 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_-_52715179 | 0.16 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr8_+_77593448 | 0.16 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr13_-_103719196 | 0.15 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr7_+_35756092 | 0.15 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr15_-_55541227 | 0.15 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr9_+_22646189 | 0.14 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr22_-_38240316 | 0.14 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr16_+_30669720 | 0.14 |
ENST00000356166.6
|
FBRS
|
fibrosin |
| chr12_+_70219052 | 0.14 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr5_-_159766528 | 0.12 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr19_+_1440838 | 0.11 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr1_-_45956868 | 0.11 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr1_+_52682052 | 0.11 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr16_-_24942411 | 0.11 |
ENST00000571843.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr1_-_45956800 | 0.11 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr17_-_10560619 | 0.10 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr16_-_24942273 | 0.10 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr4_+_68424434 | 0.10 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr2_+_173724771 | 0.10 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_68000442 | 0.09 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr17_-_73663168 | 0.08 |
ENST00000578201.1
ENST00000423245.2 |
RECQL5
|
RecQ protein-like 5 |
| chr9_+_125132803 | 0.08 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_+_48483736 | 0.08 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr1_+_84630574 | 0.08 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr10_-_94301107 | 0.07 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr6_+_122793058 | 0.07 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr21_+_30673091 | 0.07 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr10_+_104005272 | 0.07 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr4_-_69111401 | 0.06 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr16_+_2205755 | 0.06 |
ENST00000326181.6
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr1_+_84630352 | 0.05 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_78769549 | 0.05 |
ENST00000370758.1
|
PTGFR
|
prostaglandin F receptor (FP) |
| chr9_+_20927764 | 0.05 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr9_+_123884038 | 0.04 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr22_-_38240412 | 0.04 |
ENST00000215941.4
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr7_-_130353553 | 0.03 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr3_+_89156799 | 0.03 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr8_-_114449112 | 0.03 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr7_+_128784712 | 0.02 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr6_-_27782548 | 0.02 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr3_-_150690786 | 0.02 |
ENST00000327047.1
|
CLRN1
|
clarin 1 |
| chr1_-_149783914 | 0.02 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr15_+_71839566 | 0.01 |
ENST00000357769.4
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr2_-_176046391 | 0.01 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr3_-_123710199 | 0.01 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr2_+_74685413 | 0.01 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr21_+_30672433 | 0.01 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr1_-_45956822 | 0.00 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr9_-_130829588 | 0.00 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.5 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.1 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |