Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
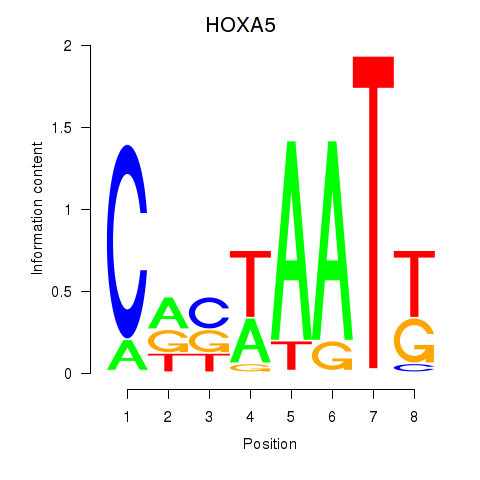
Results for HOXA5
Z-value: 0.31
Transcription factors associated with HOXA5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA5
|
ENSG00000106004.4 | homeobox A5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA5 | hg19_v2_chr7_-_27183263_27183287 | 0.52 | 4.8e-01 | Click! |
Activity profile of HOXA5 motif
Sorted Z-values of HOXA5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_99716914 | 0.19 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_-_103747011 | 0.15 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_102276192 | 0.15 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_143567262 | 0.13 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr14_+_20187174 | 0.12 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr4_-_103746924 | 0.11 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr15_+_77713222 | 0.11 |
ENST00000558176.1
|
HMG20A
|
high mobility group 20A |
| chr6_+_130339710 | 0.10 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_+_100111580 | 0.10 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr18_+_68002675 | 0.10 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr2_-_37544209 | 0.10 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr1_+_212475148 | 0.09 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr12_-_94673956 | 0.09 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr5_+_127039075 | 0.09 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr13_+_76378305 | 0.09 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chrX_+_102024075 | 0.09 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr14_+_65878650 | 0.09 |
ENST00000555559.1
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr14_+_65878565 | 0.08 |
ENST00000556518.1
ENST00000557164.1 |
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr4_-_129208030 | 0.08 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr4_-_103746683 | 0.08 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_74606223 | 0.08 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr19_-_51522955 | 0.07 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr8_+_75262629 | 0.07 |
ENST00000434412.2
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr2_-_214017151 | 0.07 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr8_-_79717750 | 0.07 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr5_+_76326187 | 0.07 |
ENST00000312916.7
ENST00000506806.1 |
AGGF1
|
angiogenic factor with G patch and FHA domains 1 |
| chr7_-_23510086 | 0.07 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr5_-_87516448 | 0.07 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr12_-_11150474 | 0.07 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr7_-_35013217 | 0.07 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr8_-_90769422 | 0.07 |
ENST00000524190.1
ENST00000523859.1 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr1_-_170043709 | 0.06 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr2_+_30670077 | 0.06 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr6_+_116575329 | 0.06 |
ENST00000430252.2
ENST00000540275.1 ENST00000448740.2 |
DSE
RP3-486I3.7
|
dermatan sulfate epimerase RP3-486I3.7 |
| chr6_+_143447322 | 0.06 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr2_+_30670127 | 0.06 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chrX_+_10124977 | 0.06 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr18_-_33709268 | 0.05 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr5_+_66300464 | 0.05 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_114163945 | 0.05 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr14_+_102276209 | 0.05 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr9_-_127905736 | 0.05 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr10_-_21186144 | 0.05 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr5_+_66300446 | 0.05 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_179322481 | 0.05 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr6_+_167525277 | 0.05 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr15_-_67351586 | 0.05 |
ENST00000558071.1
|
RP11-798K3.2
|
RP11-798K3.2 |
| chr2_-_190446738 | 0.04 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr2_-_207082748 | 0.04 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr12_+_104337515 | 0.04 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr6_+_168418553 | 0.04 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
| chr12_-_54653313 | 0.04 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr2_+_183982238 | 0.04 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr6_+_25963020 | 0.04 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr1_-_244006528 | 0.04 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr2_-_160919112 | 0.04 |
ENST00000283243.7
ENST00000392771.1 |
PLA2R1
|
phospholipase A2 receptor 1, 180kDa |
| chr2_-_61697862 | 0.04 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr12_+_111051832 | 0.04 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr3_-_78719376 | 0.04 |
ENST00000495961.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr10_-_7708918 | 0.04 |
ENST00000256861.6
ENST00000397146.2 ENST00000446830.2 ENST00000397145.2 |
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr20_-_34638841 | 0.04 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr3_+_196466710 | 0.04 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr19_-_54604083 | 0.03 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr7_+_100136811 | 0.03 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr1_-_154941350 | 0.03 |
ENST00000444179.1
ENST00000414115.1 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr10_+_126630692 | 0.03 |
ENST00000359653.4
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1 |
| chr6_-_24666819 | 0.03 |
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr3_+_37035289 | 0.03 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr15_+_77713299 | 0.03 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr16_+_202686 | 0.03 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr19_+_41768401 | 0.03 |
ENST00000352456.3
ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr14_+_102276132 | 0.03 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_-_92463210 | 0.03 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chrX_-_72097698 | 0.03 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chr2_+_227771404 | 0.03 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr15_-_73925575 | 0.03 |
ENST00000562924.1
ENST00000563691.1 ENST00000565325.1 ENST00000542234.1 |
NPTN
|
neuroplastin |
| chr14_-_20801427 | 0.03 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr7_+_26438187 | 0.03 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr2_-_40657397 | 0.03 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr16_-_66764119 | 0.03 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr4_-_113627966 | 0.03 |
ENST00000505632.1
|
RP11-148B6.2
|
RP11-148B6.2 |
| chrX_+_72062802 | 0.03 |
ENST00000373533.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr3_+_130569429 | 0.03 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr2_+_102508955 | 0.02 |
ENST00000414004.2
|
FLJ20373
|
FLJ20373 |
| chr1_-_63988846 | 0.02 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr10_-_61495760 | 0.02 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr14_+_67291158 | 0.02 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr1_+_171283331 | 0.02 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr6_+_131958436 | 0.02 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr8_-_141774467 | 0.02 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr6_-_143266297 | 0.02 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr10_+_101089107 | 0.02 |
ENST00000446890.1
ENST00000370528.3 |
CNNM1
|
cyclin M1 |
| chr1_+_27158505 | 0.02 |
ENST00000534643.1
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr11_-_96076334 | 0.02 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr3_-_79068594 | 0.02 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr9_+_70971815 | 0.02 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr1_+_84609944 | 0.02 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr20_+_58515417 | 0.02 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr6_-_32157947 | 0.02 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr10_-_115904361 | 0.02 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr4_+_1283639 | 0.02 |
ENST00000303400.4
ENST00000505177.2 ENST00000503653.1 ENST00000264750.6 ENST00000502558.1 ENST00000452175.2 ENST00000514708.1 |
MAEA
|
macrophage erythroblast attacher |
| chr1_+_160370344 | 0.02 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr1_+_153950202 | 0.02 |
ENST00000608236.1
|
RP11-422P24.11
|
RP11-422P24.11 |
| chr1_-_197169672 | 0.02 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr16_-_62070305 | 0.02 |
ENST00000584337.1
|
CDH8
|
cadherin 8, type 2 |
| chr19_-_44384291 | 0.02 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr8_+_92261516 | 0.02 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr15_-_34628951 | 0.02 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr7_-_23571586 | 0.02 |
ENST00000538367.1
ENST00000392502.4 ENST00000297071.4 |
TRA2A
|
transformer 2 alpha homolog (Drosophila) |
| chr6_-_28554977 | 0.02 |
ENST00000452236.2
|
SCAND3
|
SCAN domain containing 3 |
| chrX_+_75648046 | 0.01 |
ENST00000361470.2
|
MAGEE1
|
melanoma antigen family E, 1 |
| chr16_-_72128270 | 0.01 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr1_+_221051699 | 0.01 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr1_+_100111479 | 0.01 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr1_+_150337100 | 0.01 |
ENST00000401000.4
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr13_+_76378407 | 0.01 |
ENST00000447038.1
|
LMO7
|
LIM domain 7 |
| chr10_+_17270214 | 0.01 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr4_+_26324474 | 0.01 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_+_33061543 | 0.01 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr3_-_62359180 | 0.01 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chr20_+_4666882 | 0.01 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr6_+_125304502 | 0.01 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr4_+_86525299 | 0.01 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr7_-_36634181 | 0.01 |
ENST00000538464.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr10_+_123951957 | 0.01 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_-_41166953 | 0.01 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr10_-_52008313 | 0.01 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr11_-_115375107 | 0.01 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr6_+_155537771 | 0.01 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr12_+_111051902 | 0.01 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr4_-_164534657 | 0.01 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr6_-_152623231 | 0.01 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr18_+_13611431 | 0.01 |
ENST00000587757.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr16_-_18573396 | 0.01 |
ENST00000543392.1
ENST00000381474.3 ENST00000330537.6 |
NOMO2
|
NODAL modulator 2 |
| chr3_-_62358690 | 0.01 |
ENST00000475839.1
|
FEZF2
|
FEZ family zinc finger 2 |
| chr4_-_168155169 | 0.01 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr9_+_75766763 | 0.01 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr14_-_92413353 | 0.01 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr2_-_113594279 | 0.01 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr14_+_78174414 | 0.00 |
ENST00000557342.1
ENST00000238688.5 ENST00000557623.1 ENST00000557431.1 ENST00000556831.1 ENST00000556375.1 ENST00000553981.1 |
SLIRP
|
SRA stem-loop interacting RNA binding protein |
| chr19_-_44259053 | 0.00 |
ENST00000601170.1
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr18_+_616711 | 0.00 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr19_+_53761545 | 0.00 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr9_+_140125385 | 0.00 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr17_-_33446820 | 0.00 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr3_-_157221128 | 0.00 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_27158483 | 0.00 |
ENST00000374141.2
|
ZDHHC18
|
zinc finger, DHHC-type containing 18 |
| chr1_-_91487013 | 0.00 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.0 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.0 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.0 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |