Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
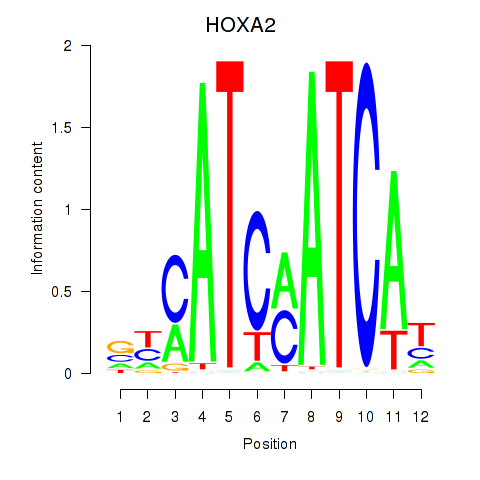
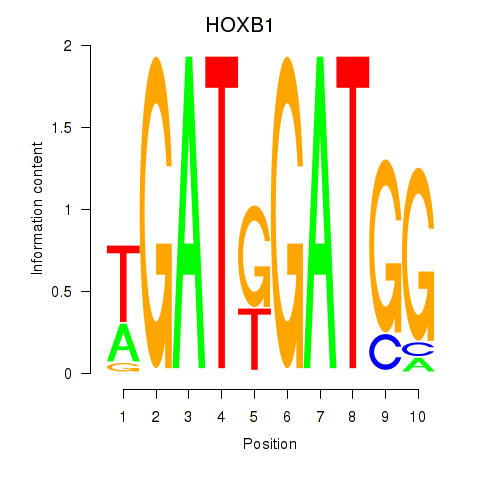
Results for HOXA2_HOXB1
Z-value: 0.23
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | -0.85 | 1.5e-01 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_159481988 | 0.25 |
ENST00000472451.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr7_-_27170352 | 0.19 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr21_-_43430440 | 0.15 |
ENST00000398505.3
ENST00000310826.5 ENST00000449949.1 ENST00000398499.1 ENST00000398497.2 ENST00000398511.3 |
ZBTB21
|
zinc finger and BTB domain containing 21 |
| chrX_+_41583408 | 0.10 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr6_+_32006042 | 0.10 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr12_+_21679220 | 0.08 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr1_-_217804377 | 0.08 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chrX_-_10645773 | 0.08 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr8_+_23386557 | 0.08 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr3_+_150264555 | 0.07 |
ENST00000406576.3
ENST00000482093.1 ENST00000273435.5 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr3_+_150264458 | 0.07 |
ENST00000487799.1
ENST00000460851.1 |
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa |
| chr16_+_85936295 | 0.07 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr5_+_162864575 | 0.07 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr8_+_143530791 | 0.07 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr2_+_12857043 | 0.06 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr17_-_57229155 | 0.06 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr6_-_159065741 | 0.06 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chrX_+_95939638 | 0.06 |
ENST00000373061.3
ENST00000373054.4 ENST00000355827.4 |
DIAPH2
|
diaphanous-related formin 2 |
| chr12_+_10658489 | 0.06 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr1_-_92351666 | 0.06 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr10_+_24755416 | 0.05 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr2_+_135011731 | 0.05 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr2_-_200322723 | 0.05 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr6_-_64029879 | 0.05 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr7_-_100881041 | 0.05 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr5_+_98109322 | 0.05 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr10_+_102505468 | 0.05 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chr10_+_89264625 | 0.05 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr11_+_57365150 | 0.05 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr5_+_145826867 | 0.05 |
ENST00000296702.5
ENST00000394421.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr7_-_143956815 | 0.04 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr8_+_23386305 | 0.04 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr2_-_200323414 | 0.04 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr9_-_15250114 | 0.04 |
ENST00000507993.1
|
TTC39B
|
tetratricopeptide repeat domain 39B |
| chr12_+_21168630 | 0.04 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr1_-_67390474 | 0.04 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr16_-_11617444 | 0.04 |
ENST00000598234.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr4_-_90757364 | 0.04 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_52082751 | 0.04 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr8_-_101730061 | 0.03 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_+_7022909 | 0.03 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr2_-_190044480 | 0.03 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr11_-_126174186 | 0.03 |
ENST00000524964.1
|
RP11-712L6.5
|
Uncharacterized protein |
| chr3_-_150264272 | 0.03 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr19_+_13051206 | 0.03 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr19_-_41356347 | 0.03 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr19_-_45735138 | 0.03 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr10_-_79397547 | 0.03 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_21608525 | 0.03 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr16_+_85645007 | 0.03 |
ENST00000405402.2
|
GSE1
|
Gse1 coiled-coil protein |
| chrX_-_30993201 | 0.03 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr8_-_79717750 | 0.03 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr2_+_176995011 | 0.03 |
ENST00000548663.1
ENST00000450510.2 |
HOXD8
|
homeobox D8 |
| chr16_+_66429358 | 0.03 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr7_-_27135591 | 0.03 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr11_+_133902547 | 0.03 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr2_+_54684327 | 0.03 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr1_+_67390578 | 0.02 |
ENST00000371018.3
ENST00000355977.6 ENST00000357692.2 ENST00000401041.1 ENST00000371016.1 ENST00000371014.1 ENST00000371012.2 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr3_+_239652 | 0.02 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr4_-_90756769 | 0.02 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_-_49944806 | 0.02 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr1_-_109584608 | 0.02 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr5_-_179045199 | 0.02 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr16_-_49890016 | 0.02 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr9_+_132099158 | 0.02 |
ENST00000444125.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr17_-_42277203 | 0.02 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr20_+_34742650 | 0.02 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr10_-_105615164 | 0.02 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr20_+_42574317 | 0.02 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr9_+_40028620 | 0.02 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr2_+_114647504 | 0.02 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chrX_+_95939711 | 0.02 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr7_-_86595190 | 0.02 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr12_-_86650077 | 0.02 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_+_18943554 | 0.02 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr17_+_79495397 | 0.02 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr1_+_68150744 | 0.02 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr1_-_92351769 | 0.02 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr11_-_2162162 | 0.02 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_-_156877997 | 0.02 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr1_+_178062855 | 0.02 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr12_+_101988627 | 0.01 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr1_-_109584716 | 0.01 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr6_+_33387868 | 0.01 |
ENST00000418600.2
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1 |
| chr19_-_12997995 | 0.01 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr7_+_86273218 | 0.01 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr1_-_109584768 | 0.01 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr2_+_242089833 | 0.01 |
ENST00000404405.3
ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr11_-_126870655 | 0.01 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr13_+_97928395 | 0.01 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_167181917 | 0.01 |
ENST00000519204.1
|
TENM2
|
teneurin transmembrane protein 2 |
| chr1_-_223536679 | 0.01 |
ENST00000608996.1
|
SUSD4
|
sushi domain containing 4 |
| chr17_-_37353950 | 0.01 |
ENST00000394310.3
ENST00000394303.3 ENST00000344140.5 |
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr12_+_7282795 | 0.01 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr2_-_197226875 | 0.01 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr3_-_98619999 | 0.01 |
ENST00000449482.1
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2 |
| chr10_-_65028938 | 0.01 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr14_-_54425475 | 0.01 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr12_+_101988774 | 0.01 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr15_+_81591757 | 0.01 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr17_+_1173853 | 0.01 |
ENST00000391429.1
|
BHLHA9
|
basic helix-loop-helix family, member a9 |
| chr15_-_74501310 | 0.01 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr6_+_32006159 | 0.01 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr12_-_118490217 | 0.01 |
ENST00000542304.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr16_+_15596123 | 0.01 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr17_+_58755184 | 0.01 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr9_-_73029540 | 0.01 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr6_-_113754604 | 0.01 |
ENST00000421737.1
|
RP1-124C6.1
|
RP1-124C6.1 |
| chr7_+_79765071 | 0.01 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr12_+_57482877 | 0.01 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr1_+_243419306 | 0.01 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr19_-_51512804 | 0.01 |
ENST00000594211.1
ENST00000376832.4 |
KLK9
|
kallikrein-related peptidase 9 |
| chr20_+_30467600 | 0.01 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr14_-_24733444 | 0.00 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr3_+_191046810 | 0.00 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr2_-_44588694 | 0.00 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr12_+_57610562 | 0.00 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr9_-_101471479 | 0.00 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr15_-_64673630 | 0.00 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr2_-_44588679 | 0.00 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr13_+_52586517 | 0.00 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr8_+_22250334 | 0.00 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_+_26624775 | 0.00 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr11_-_93583697 | 0.00 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr1_-_65533390 | 0.00 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr10_+_95848824 | 0.00 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chrX_-_110655306 | 0.00 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr15_+_48009541 | 0.00 |
ENST00000536845.2
ENST00000558816.1 |
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr15_-_74501360 | 0.00 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr2_-_145277882 | 0.00 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0060979 | transforming growth factor beta receptor complex assembly(GO:0007181) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.0 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.0 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |