Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HOXA10_HOXB9
Z-value: 0.56
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
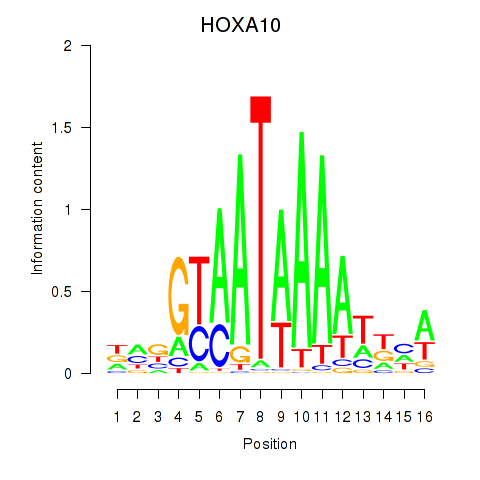
HOXA10
|
ENSG00000253293.3 | homeobox A10 |
|
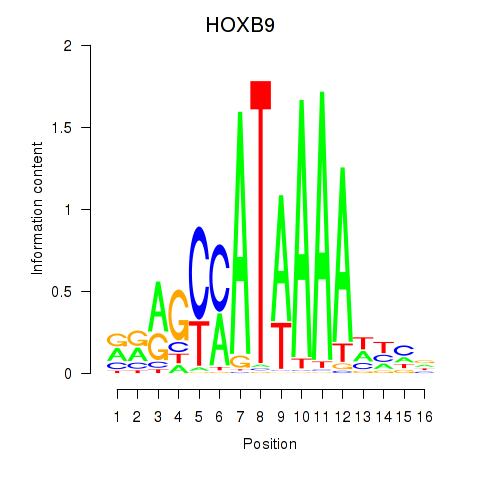
HOXB9
|
ENSG00000170689.8 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27219849_27219880 | 0.50 | 5.0e-01 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | -0.37 | 6.3e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_91152303 | 0.67 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr6_+_32812568 | 0.66 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr7_-_41742697 | 0.66 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr2_-_216878305 | 0.49 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr4_-_76957214 | 0.44 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr17_-_39646116 | 0.44 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr7_+_35756186 | 0.42 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr4_-_129491686 | 0.38 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr11_-_104905840 | 0.34 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr2_+_101591314 | 0.32 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr22_-_31324215 | 0.31 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr3_-_172241250 | 0.31 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_-_104916034 | 0.30 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr11_-_104972158 | 0.30 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr15_+_67418047 | 0.29 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr15_-_101817492 | 0.29 |
ENST00000528346.1
ENST00000531964.1 |
VIMP
|
VCP-interacting membrane protein |
| chr14_-_25479811 | 0.29 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr4_-_140222358 | 0.28 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr10_+_102106829 | 0.28 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr7_+_35756092 | 0.28 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr5_-_126409159 | 0.26 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr2_-_32390801 | 0.26 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr15_-_55541227 | 0.26 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr22_+_43011247 | 0.23 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr13_-_103719196 | 0.23 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr4_-_72649763 | 0.22 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr10_+_78078088 | 0.21 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr19_+_4402659 | 0.21 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr1_+_104615595 | 0.21 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr1_+_23345943 | 0.21 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr4_+_70894130 | 0.20 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr10_-_73497581 | 0.20 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr6_+_46761118 | 0.19 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr17_+_61086917 | 0.18 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_+_26156551 | 0.18 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr9_-_133814527 | 0.18 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chrX_-_133792480 | 0.18 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr8_+_145215928 | 0.18 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr15_+_101417919 | 0.18 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr1_+_209878182 | 0.18 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr3_+_157828152 | 0.17 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr6_+_28249299 | 0.17 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_+_28249332 | 0.17 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr9_+_137979506 | 0.17 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr18_+_3252206 | 0.17 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_+_23345930 | 0.17 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr1_+_52682052 | 0.17 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr5_-_150467221 | 0.17 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_+_74701062 | 0.16 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_-_155880672 | 0.16 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr1_-_23521222 | 0.15 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr18_+_3252265 | 0.15 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_26250835 | 0.15 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr4_-_71532601 | 0.15 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr22_-_38240316 | 0.15 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr10_-_99030395 | 0.14 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr18_+_19192228 | 0.14 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr20_-_33732952 | 0.14 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr2_+_181988560 | 0.14 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr10_-_73848531 | 0.14 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chrX_-_15619076 | 0.14 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr1_-_23520755 | 0.14 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr8_+_77593448 | 0.14 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr15_-_59500973 | 0.14 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr12_+_1800179 | 0.13 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr11_-_65359947 | 0.13 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr9_+_134065506 | 0.13 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr9_-_130889990 | 0.13 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr12_+_4130143 | 0.13 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr17_-_38928414 | 0.12 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr15_+_57891609 | 0.12 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr18_-_52989525 | 0.12 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr10_-_73848086 | 0.12 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr15_+_75639372 | 0.12 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr4_-_19458597 | 0.12 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr12_+_27623565 | 0.12 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr5_-_58295712 | 0.12 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr9_+_34652164 | 0.12 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr2_-_154335300 | 0.11 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr19_+_3185910 | 0.11 |
ENST00000588428.1
|
NCLN
|
nicalin |
| chr7_-_7782204 | 0.11 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr6_+_12717892 | 0.11 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr20_+_3767547 | 0.11 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr8_+_21906433 | 0.11 |
ENST00000522148.1
|
DMTN
|
dematin actin binding protein |
| chr16_-_12070468 | 0.11 |
ENST00000538896.1
|
RP11-166B2.1
|
Putative NPIP-like protein LOC729978 |
| chr4_-_71532207 | 0.11 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr22_-_23922410 | 0.11 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr7_-_122840015 | 0.10 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr7_+_141408153 | 0.10 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr22_-_23922448 | 0.10 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr5_-_139937895 | 0.10 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr10_+_47658234 | 0.10 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr4_+_68424434 | 0.10 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr19_-_46580352 | 0.10 |
ENST00000601672.1
|
IGFL4
|
IGF-like family member 4 |
| chr1_-_161207953 | 0.10 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr11_+_61717279 | 0.10 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr1_+_160370344 | 0.10 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr9_-_133814455 | 0.10 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr7_-_50860565 | 0.10 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr9_+_105757590 | 0.10 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr19_-_3500635 | 0.09 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr4_+_146539415 | 0.09 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr18_+_68002675 | 0.09 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr4_+_129349188 | 0.09 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr11_-_58611957 | 0.09 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr9_+_82187487 | 0.09 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_-_158182322 | 0.09 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr7_+_128784712 | 0.09 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr14_+_73706308 | 0.09 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr3_+_107364769 | 0.09 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr2_+_208423891 | 0.09 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chrX_-_54070388 | 0.09 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr5_+_158737824 | 0.09 |
ENST00000521472.1
|
AC008697.1
|
AC008697.1 |
| chr9_+_82187630 | 0.08 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_+_234668894 | 0.08 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_-_186430222 | 0.08 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr16_-_25122735 | 0.08 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr1_-_161208013 | 0.08 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr9_+_70856397 | 0.08 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr5_-_159766528 | 0.08 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr11_-_104827425 | 0.08 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chrX_+_36053908 | 0.08 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr1_-_94147385 | 0.08 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr19_-_29704448 | 0.08 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr9_-_79307096 | 0.08 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr12_-_118628315 | 0.08 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chrY_-_20935572 | 0.08 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr2_-_158182105 | 0.08 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chrX_-_100872911 | 0.07 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chrX_+_36254051 | 0.07 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr8_+_132952112 | 0.07 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr4_+_79472975 | 0.07 |
ENST00000512373.1
ENST00000514171.1 |
ANXA3
|
annexin A3 |
| chr8_+_1993152 | 0.07 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr11_+_10471836 | 0.07 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr19_-_10420459 | 0.07 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr2_+_220144052 | 0.07 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr10_-_94301107 | 0.07 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr12_-_21928515 | 0.07 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr2_+_20101786 | 0.07 |
ENST00000607190.1
|
RP11-79O8.1
|
RP11-79O8.1 |
| chr20_-_25320367 | 0.07 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chrY_+_20708557 | 0.07 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr12_+_12510352 | 0.07 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr4_-_71532668 | 0.07 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr6_+_111408698 | 0.07 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr1_+_84630574 | 0.07 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_+_56584414 | 0.07 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr2_+_220143989 | 0.07 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr3_+_107602030 | 0.06 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr15_+_75639296 | 0.06 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr12_+_12938541 | 0.06 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr17_-_67264947 | 0.06 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr4_-_170897045 | 0.06 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr14_-_38028689 | 0.06 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chrX_+_65384052 | 0.06 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr21_+_30503282 | 0.06 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr22_-_22292934 | 0.06 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr1_-_161207986 | 0.06 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chrX_-_110655306 | 0.06 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr7_+_18536090 | 0.06 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr4_-_69817481 | 0.06 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr22_-_17302589 | 0.06 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr8_+_66955648 | 0.06 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr14_+_79745746 | 0.06 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr8_-_21669826 | 0.06 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr6_+_53948328 | 0.06 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr8_+_1993173 | 0.06 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr2_+_102928009 | 0.06 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr15_+_48483736 | 0.06 |
ENST00000417307.2
ENST00000559641.1 |
CTXN2
SLC12A1
|
cortexin 2 solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chrX_+_36246735 | 0.06 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr16_-_48281444 | 0.06 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr17_-_73663245 | 0.06 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr9_+_77112244 | 0.06 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chrX_-_73072534 | 0.06 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr15_-_50838888 | 0.06 |
ENST00000532404.1
|
USP50
|
ubiquitin specific peptidase 50 |
| chr11_+_22688150 | 0.06 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr16_-_21663950 | 0.06 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr14_+_35514323 | 0.05 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr16_-_30621663 | 0.05 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr11_-_46113756 | 0.05 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr4_+_88896819 | 0.05 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr1_-_143767881 | 0.05 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr22_-_43010928 | 0.05 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_-_75498553 | 0.05 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr19_+_12175504 | 0.05 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr3_+_57875711 | 0.05 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr18_+_22040620 | 0.05 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr14_+_39944025 | 0.05 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr4_+_169575875 | 0.05 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_+_128426535 | 0.05 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr3_-_18480260 | 0.05 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr3_-_38992052 | 0.05 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr11_-_5537920 | 0.05 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr2_-_122262593 | 0.05 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr19_+_15838834 | 0.05 |
ENST00000305899.3
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2 |
| chr6_-_166582107 | 0.05 |
ENST00000296946.2
ENST00000461348.2 ENST00000366871.3 |
T
|
T, brachyury homolog (mouse) |
| chr3_+_125687987 | 0.05 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr15_+_40886439 | 0.05 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr7_-_78400598 | 0.05 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_-_10560619 | 0.05 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr9_+_125132803 | 0.05 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 0.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.2 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0090196 | positive regulation of macrophage activation(GO:0043032) chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0003051 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:2000861 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.9 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.0 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.4 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |