Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
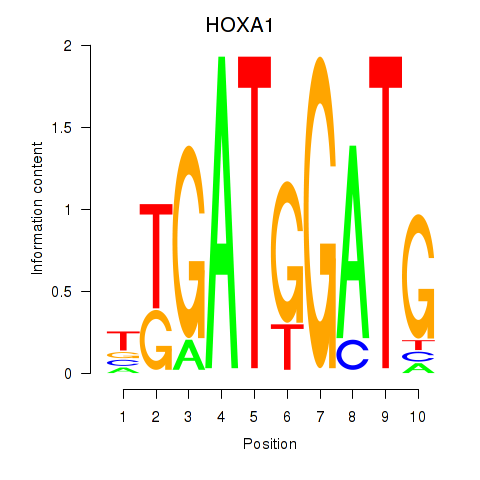
Results for HOXA1
Z-value: 0.79
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.7 | homeobox A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg19_v2_chr7_-_27135591_27135658 | -0.92 | 8.4e-02 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10697895 | 1.56 |
ENST00000591240.1
ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr8_-_22549856 | 0.52 |
ENST00000522910.1
|
EGR3
|
early growth response 3 |
| chr5_+_162864575 | 0.46 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr20_-_45976816 | 0.43 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr19_+_16435625 | 0.38 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr12_+_69742121 | 0.37 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chrX_-_63005405 | 0.37 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr15_-_64673630 | 0.34 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr15_-_93616340 | 0.33 |
ENST00000557420.1
ENST00000542321.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr15_-_64673665 | 0.28 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr17_+_26800296 | 0.26 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr7_-_120497178 | 0.26 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr9_-_116861337 | 0.25 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr6_+_80129989 | 0.25 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr17_-_79359046 | 0.24 |
ENST00000574041.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr5_-_125930929 | 0.23 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr11_-_111649015 | 0.23 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr1_-_151319710 | 0.23 |
ENST00000290524.4
ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr17_+_40932610 | 0.22 |
ENST00000246914.5
|
WNK4
|
WNK lysine deficient protein kinase 4 |
| chr8_-_42360015 | 0.21 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr9_-_112970436 | 0.21 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr12_-_71533055 | 0.21 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chrX_+_154610428 | 0.20 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
| chr16_-_20566616 | 0.20 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr2_-_75788428 | 0.19 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr8_-_27468842 | 0.19 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr11_-_93583697 | 0.19 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr11_+_2482661 | 0.19 |
ENST00000335475.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr8_-_27468945 | 0.18 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr19_-_39368887 | 0.18 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr19_-_16008880 | 0.18 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr5_-_125930877 | 0.17 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr3_+_196366616 | 0.17 |
ENST00000426755.1
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr16_-_11477562 | 0.17 |
ENST00000595168.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr10_+_62538248 | 0.17 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr11_-_104034827 | 0.17 |
ENST00000393158.2
|
PDGFD
|
platelet derived growth factor D |
| chr1_-_20306909 | 0.17 |
ENST00000375111.3
ENST00000400520.3 |
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr22_+_41968007 | 0.17 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr8_+_104152922 | 0.16 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr12_-_89919965 | 0.16 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr10_-_102046098 | 0.16 |
ENST00000441611.1
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr8_-_27469196 | 0.15 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr17_-_57229155 | 0.15 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr16_-_28303360 | 0.15 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr11_-_104035088 | 0.15 |
ENST00000302251.5
|
PDGFD
|
platelet derived growth factor D |
| chr2_+_65215604 | 0.14 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr1_+_85527987 | 0.14 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr7_+_74379083 | 0.14 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr5_-_42825983 | 0.14 |
ENST00000506577.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr1_-_45140074 | 0.13 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr19_+_19144384 | 0.13 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr4_-_38806404 | 0.13 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr7_+_139528952 | 0.13 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr1_-_207095324 | 0.13 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr7_+_139529040 | 0.13 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr17_+_43239231 | 0.13 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr17_+_43239191 | 0.13 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr10_+_62538089 | 0.13 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr19_-_33360647 | 0.13 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr14_-_81408063 | 0.12 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr13_-_52026730 | 0.12 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr11_+_63870660 | 0.12 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr14_+_21569245 | 0.12 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr17_+_68071389 | 0.12 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_71774516 | 0.12 |
ENST00000425534.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chrX_+_70443050 | 0.12 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr1_+_226250379 | 0.12 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chr2_-_75788424 | 0.11 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr17_-_3499125 | 0.11 |
ENST00000399759.3
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr18_-_74207146 | 0.11 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr5_+_36152163 | 0.11 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chrX_+_37639302 | 0.11 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr5_-_173173171 | 0.11 |
ENST00000517733.1
ENST00000518902.1 |
CTB-43E15.3
|
CTB-43E15.3 |
| chr5_+_133861790 | 0.11 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr8_-_27468717 | 0.11 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr1_-_53018654 | 0.11 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr2_-_44588694 | 0.11 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr9_-_128246769 | 0.10 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr5_+_159656437 | 0.10 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr19_-_42192096 | 0.10 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr17_-_79359021 | 0.10 |
ENST00000572301.1
|
RP11-1055B8.3
|
RP11-1055B8.3 |
| chr4_-_144826682 | 0.10 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr12_+_75728419 | 0.10 |
ENST00000378695.4
ENST00000312442.2 |
GLIPR1L1
|
GLI pathogenesis-related 1 like 1 |
| chr1_-_201096312 | 0.10 |
ENST00000449188.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr2_-_225434538 | 0.10 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr6_-_31689456 | 0.10 |
ENST00000495859.1
ENST00000375819.2 |
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C |
| chr14_+_24779340 | 0.10 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr2_+_161993465 | 0.10 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_94139105 | 0.10 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr2_-_44588679 | 0.10 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr20_+_34742650 | 0.10 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr3_-_149093499 | 0.10 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr11_+_34460447 | 0.10 |
ENST00000241052.4
|
CAT
|
catalase |
| chr5_-_693500 | 0.10 |
ENST00000360578.5
|
TPPP
|
tubulin polymerization promoting protein |
| chr12_-_95510743 | 0.09 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr15_-_93616892 | 0.09 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr1_-_182369751 | 0.09 |
ENST00000367565.1
|
TEDDM1
|
transmembrane epididymal protein 1 |
| chr8_+_22132847 | 0.09 |
ENST00000521356.1
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr1_+_165864800 | 0.09 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr15_+_84908573 | 0.09 |
ENST00000424966.1
ENST00000422563.2 |
GOLGA6L4
|
golgin A6 family-like 4 |
| chr9_+_33795533 | 0.09 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr21_-_46340884 | 0.09 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr12_-_110906027 | 0.09 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr10_-_105615164 | 0.09 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chrX_-_154493791 | 0.09 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr7_+_45067265 | 0.09 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr18_+_11752040 | 0.09 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr22_-_39190116 | 0.09 |
ENST00000406622.1
ENST00000216068.4 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr8_-_79717163 | 0.09 |
ENST00000520269.1
|
IL7
|
interleukin 7 |
| chr17_-_44896047 | 0.08 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr16_+_28303804 | 0.08 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr20_-_30458491 | 0.08 |
ENST00000339738.5
|
DUSP15
|
dual specificity phosphatase 15 |
| chr5_+_133861339 | 0.08 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr1_-_153517473 | 0.08 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr3_-_129375556 | 0.08 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr1_+_162602244 | 0.08 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr2_+_87808725 | 0.08 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr9_-_5304432 | 0.08 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr7_-_27170352 | 0.08 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr13_-_20077417 | 0.08 |
ENST00000382978.1
ENST00000400230.2 ENST00000255310.6 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr12_+_65996599 | 0.07 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr22_-_22641542 | 0.07 |
ENST00000606654.1
|
LL22NC03-2H8.5
|
LL22NC03-2H8.5 |
| chr16_-_66584015 | 0.07 |
ENST00000545043.2
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr16_+_75256507 | 0.07 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr3_-_179322416 | 0.07 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr12_+_64173583 | 0.07 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr1_-_33338076 | 0.07 |
ENST00000496770.1
|
FNDC5
|
fibronectin type III domain containing 5 |
| chr8_-_27462822 | 0.07 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr5_-_141338627 | 0.07 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr1_-_211666259 | 0.07 |
ENST00000367002.4
|
RD3
|
retinal degeneration 3 |
| chr17_+_39975455 | 0.07 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr4_+_71600063 | 0.07 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr4_+_2451700 | 0.07 |
ENST00000506607.1
|
RP11-503N18.3
|
Uncharacterized protein |
| chr17_+_39975544 | 0.07 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr11_+_46193466 | 0.07 |
ENST00000533793.1
|
RP11-702F3.3
|
RP11-702F3.3 |
| chr15_-_75134992 | 0.07 |
ENST00000568667.1
|
ULK3
|
unc-51 like kinase 3 |
| chr19_-_15544099 | 0.06 |
ENST00000599910.2
|
WIZ
|
widely interspaced zinc finger motifs |
| chr13_-_20735178 | 0.06 |
ENST00000241125.3
|
GJA3
|
gap junction protein, alpha 3, 46kDa |
| chr10_+_98741041 | 0.06 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr20_-_62168714 | 0.06 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr20_+_60174827 | 0.06 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr10_+_99349450 | 0.06 |
ENST00000370640.3
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr7_+_20370746 | 0.06 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chrX_-_154060946 | 0.06 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chr3_-_179322436 | 0.06 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr19_+_38880695 | 0.06 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr1_-_39347255 | 0.06 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr12_+_48577366 | 0.06 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68 |
| chr12_+_125478241 | 0.06 |
ENST00000341446.8
|
BRI3BP
|
BRI3 binding protein |
| chr7_+_30174668 | 0.06 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr12_+_8276433 | 0.06 |
ENST00000345999.3
ENST00000352620.3 ENST00000360500.3 |
CLEC4A
|
C-type lectin domain family 4, member A |
| chr2_-_170681375 | 0.06 |
ENST00000410097.1
ENST00000308099.3 ENST00000409837.1 ENST00000538491.1 ENST00000260953.5 ENST00000409965.1 ENST00000392640.2 |
METTL5
|
methyltransferase like 5 |
| chr8_-_22550691 | 0.05 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr16_-_21863541 | 0.05 |
ENST00000543654.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr12_+_21525818 | 0.05 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr1_-_13673511 | 0.05 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr9_+_124329336 | 0.05 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr13_-_24895566 | 0.05 |
ENST00000422229.2
|
AL359736.1
|
protein PCOTH isoform 1 |
| chr2_-_170681324 | 0.05 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chr12_-_10978957 | 0.05 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr10_-_98945677 | 0.05 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr1_-_45140227 | 0.05 |
ENST00000372237.3
|
TMEM53
|
transmembrane protein 53 |
| chr14_+_57046500 | 0.05 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr2_+_264869 | 0.05 |
ENST00000272067.6
ENST00000272065.5 ENST00000407983.3 |
ACP1
|
acid phosphatase 1, soluble |
| chr2_-_112237835 | 0.05 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr3_+_48264816 | 0.05 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chr20_-_30458354 | 0.05 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr2_+_44396000 | 0.05 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr16_+_28985542 | 0.05 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr19_+_58193337 | 0.05 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr12_+_57610562 | 0.05 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr12_+_57998400 | 0.05 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr6_-_47009996 | 0.05 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr10_-_75012360 | 0.05 |
ENST00000416782.2
ENST00000372945.3 ENST00000372940.3 |
MRPS16
|
mitochondrial ribosomal protein S16 |
| chr19_+_58193388 | 0.05 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr7_+_30174574 | 0.05 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_+_68150744 | 0.04 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr22_+_25615489 | 0.04 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr14_+_24702127 | 0.04 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr12_+_54402790 | 0.04 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr11_+_66247478 | 0.04 |
ENST00000531863.1
ENST00000532677.1 |
DPP3
|
dipeptidyl-peptidase 3 |
| chr9_+_10613163 | 0.04 |
ENST00000429581.2
|
RP11-87N24.2
|
RP11-87N24.2 |
| chr9_+_2717502 | 0.04 |
ENST00000382082.3
|
KCNV2
|
potassium channel, subfamily V, member 2 |
| chr12_-_96794330 | 0.04 |
ENST00000261211.3
|
CDK17
|
cyclin-dependent kinase 17 |
| chr7_-_126892303 | 0.04 |
ENST00000358373.3
|
GRM8
|
glutamate receptor, metabotropic 8 |
| chr20_+_8112824 | 0.04 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr16_+_53920795 | 0.04 |
ENST00000431610.2
ENST00000460382.1 |
FTO
|
fat mass and obesity associated |
| chr5_-_36152031 | 0.04 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr3_-_49967292 | 0.04 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr5_+_177433406 | 0.04 |
ENST00000504530.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr14_+_96949319 | 0.04 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr11_+_5509915 | 0.04 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1 |
| chr17_-_38821373 | 0.04 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_+_45140360 | 0.04 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr9_+_35829208 | 0.04 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr2_-_158345341 | 0.04 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr14_+_24779376 | 0.04 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chrX_+_9431324 | 0.04 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr16_+_29674540 | 0.04 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr15_+_101402041 | 0.04 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 0.4 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.7 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.4 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.3 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 1.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.3 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |