Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
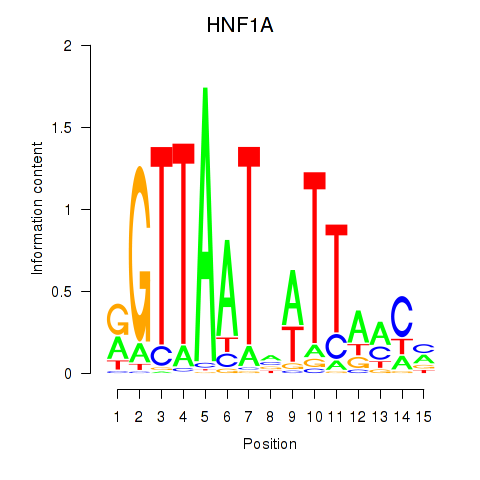
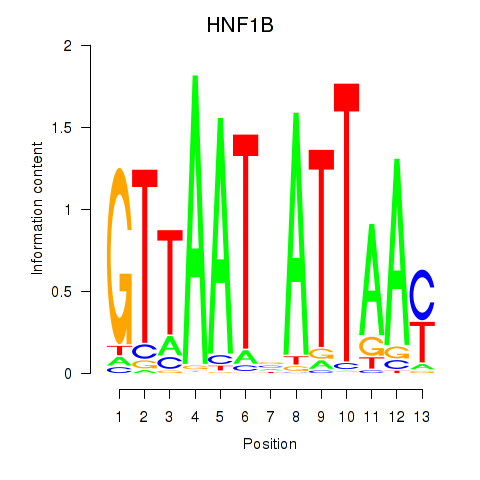
Results for HNF1A_HNF1B
Z-value: 5.56
Transcription factors associated with HNF1A_HNF1B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF1A
|
ENSG00000135100.13 | HNF1 homeobox A |
|
HNF1B
|
ENSG00000108753.8 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF1A | hg19_v2_chr12_+_121416437_121416479 | 0.98 | 2.4e-02 | Click! |
| HNF1B | hg19_v2_chr17_-_36105009_36105060 | -0.00 | 1.0e+00 | Click! |
Activity profile of HNF1A_HNF1B motif
Sorted Z-values of HNF1A_HNF1B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_64225508 | 12.50 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr19_-_36304201 | 11.32 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_176514413 | 9.25 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr9_-_123812542 | 8.90 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr10_+_115312766 | 8.12 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr4_+_155484103 | 7.66 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr1_+_48688357 | 7.57 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr1_-_197036364 | 7.56 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr2_+_38177575 | 7.40 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr4_+_3443614 | 7.29 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr4_+_155484155 | 7.15 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr10_+_99344071 | 7.05 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr9_-_116861337 | 6.66 |
ENST00000374118.3
|
KIF12
|
kinesin family member 12 |
| chr5_+_176513895 | 6.60 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr7_-_50633078 | 6.45 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr16_+_20775024 | 6.37 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr8_-_95220775 | 6.18 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr19_+_49467232 | 5.96 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr4_-_155511887 | 5.81 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr20_-_7921090 | 5.77 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr1_+_207277632 | 5.56 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr2_-_228244013 | 5.50 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr1_+_18807424 | 5.43 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr16_-_66952779 | 5.36 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr5_+_176513868 | 5.27 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr4_-_110723335 | 5.19 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr1_+_207277590 | 5.07 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr16_-_66952742 | 4.87 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr10_+_115312825 | 4.64 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr4_+_141264597 | 4.33 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr4_-_110723194 | 4.23 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr20_+_43029911 | 4.18 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr16_+_21244986 | 4.16 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr4_-_110723134 | 4.15 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr8_-_17752996 | 4.13 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr16_+_20775358 | 3.97 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr10_+_101542462 | 3.94 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr5_-_138718973 | 3.88 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr8_-_124749609 | 3.83 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr11_-_64660916 | 3.72 |
ENST00000413053.1
|
MIR194-2
|
microRNA 194-2 |
| chr16_-_69385681 | 3.50 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr2_-_47572105 | 3.05 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr14_-_94759595 | 2.92 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr11_-_111649015 | 2.91 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr10_+_99344104 | 2.85 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr17_+_73642486 | 2.76 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr12_-_39837192 | 2.64 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr14_-_94759408 | 2.49 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759361 | 2.48 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr19_+_50016610 | 2.31 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr14_+_65170820 | 2.30 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr6_-_52628271 | 2.17 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr17_+_73642315 | 2.15 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr12_+_100867486 | 2.01 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr14_-_94854926 | 1.98 |
ENST00000402629.1
ENST00000556091.1 ENST00000554720.1 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr1_-_207206092 | 1.94 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr14_+_21156915 | 1.94 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr1_-_229406746 | 1.85 |
ENST00000429227.1
ENST00000436334.1 |
RP5-1061H20.4
|
RP5-1061H20.4 |
| chr19_-_49565254 | 1.84 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr19_+_50016411 | 1.83 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr20_+_42187682 | 1.80 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr9_-_215744 | 1.75 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr16_+_89696692 | 1.59 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr11_-_117695449 | 1.58 |
ENST00000292079.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr17_+_26800296 | 1.56 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr4_-_987164 | 1.46 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr17_+_68071389 | 1.42 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_-_76944621 | 1.39 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_+_161123270 | 1.29 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr1_+_84767289 | 1.28 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr17_+_1646130 | 1.22 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr4_+_124571409 | 1.20 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr7_-_17598506 | 1.13 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr16_-_75284758 | 1.09 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr12_+_100867694 | 1.09 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr22_-_32651326 | 1.08 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr7_-_155601766 | 1.06 |
ENST00000430104.1
|
SHH
|
sonic hedgehog |
| chr20_+_42984330 | 1.05 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr6_-_25874440 | 1.01 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr11_+_560956 | 1.00 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr22_+_45098067 | 0.98 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr17_+_26800648 | 0.94 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr4_-_987217 | 0.93 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr22_-_38506619 | 0.92 |
ENST00000332536.5
ENST00000381669.3 |
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr2_+_234637754 | 0.91 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_+_26800756 | 0.91 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr12_+_100867733 | 0.88 |
ENST00000546380.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr12_+_100897130 | 0.87 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_+_229406847 | 0.82 |
ENST00000366690.4
|
RAB4A
|
RAB4A, member RAS oncogene family |
| chr7_+_152456904 | 0.82 |
ENST00000537264.1
|
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr14_-_65409502 | 0.78 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr7_+_141695633 | 0.78 |
ENST00000549489.2
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr14_+_39703084 | 0.76 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr7_+_152456829 | 0.76 |
ENST00000377776.3
ENST00000256001.8 ENST00000397282.2 |
ACTR3B
|
ARP3 actin-related protein 3 homolog B (yeast) |
| chr9_+_140125385 | 0.76 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr12_-_56615485 | 0.75 |
ENST00000549038.1
ENST00000552244.1 |
RNF41
|
ring finger protein 41 |
| chr17_+_48823975 | 0.74 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr13_+_96085847 | 0.74 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr13_-_41593425 | 0.74 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr2_+_191792376 | 0.73 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr22_+_31002990 | 0.71 |
ENST00000423350.1
|
TCN2
|
transcobalamin II |
| chr5_+_68860949 | 0.70 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr17_+_68071458 | 0.69 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr16_-_87970122 | 0.68 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chr1_-_114430169 | 0.67 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr4_-_69536346 | 0.66 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr14_-_65409438 | 0.65 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr4_-_100212132 | 0.65 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr10_-_75193308 | 0.64 |
ENST00000299432.2
|
MSS51
|
MSS51 mitochondrial translational activator |
| chr4_-_109541539 | 0.64 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr4_-_1166954 | 0.63 |
ENST00000514490.1
ENST00000431380.1 ENST00000503765.1 |
SPON2
|
spondin 2, extracellular matrix protein |
| chr4_-_166034029 | 0.63 |
ENST00000306480.6
|
TMEM192
|
transmembrane protein 192 |
| chr7_+_130126165 | 0.63 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr9_+_140125209 | 0.63 |
ENST00000538474.1
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr7_+_141695671 | 0.62 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr11_+_46740730 | 0.58 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr18_-_70931689 | 0.58 |
ENST00000581862.1
|
RP11-169F17.1
|
Protein LOC400655 |
| chr4_-_102267953 | 0.58 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_+_45406627 | 0.56 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr10_-_38146510 | 0.56 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr11_-_117170403 | 0.55 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr10_+_74451883 | 0.55 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr10_-_75226166 | 0.55 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr19_+_6464243 | 0.53 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chrX_+_120181457 | 0.52 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr6_-_113953705 | 0.51 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr3_+_107318157 | 0.50 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr19_-_43099070 | 0.50 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr7_+_130126012 | 0.50 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr10_-_52645416 | 0.49 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr21_-_35340759 | 0.48 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr17_+_41363854 | 0.47 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr19_-_57678811 | 0.46 |
ENST00000554048.2
|
DUXA
|
double homeobox A |
| chr10_-_38146482 | 0.45 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248 |
| chr16_+_22524379 | 0.43 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr19_-_38916839 | 0.43 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr10_-_101690650 | 0.43 |
ENST00000543621.1
|
DNMBP
|
dynamin binding protein |
| chr14_+_65171315 | 0.40 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr8_+_55047763 | 0.38 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr22_-_22337204 | 0.38 |
ENST00000430142.1
ENST00000357179.5 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr12_+_20963632 | 0.38 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr7_-_47579188 | 0.37 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr12_+_20963647 | 0.36 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr2_+_175199674 | 0.35 |
ENST00000394967.2
|
SP9
|
Sp9 transcription factor |
| chr2_+_234627424 | 0.35 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr3_-_107596910 | 0.34 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr3_-_192445289 | 0.33 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr20_+_42187608 | 0.33 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr2_+_28718921 | 0.31 |
ENST00000327757.5
ENST00000422425.2 ENST00000404858.1 |
PLB1
|
phospholipase B1 |
| chr1_+_104159999 | 0.31 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr2_+_234621551 | 0.30 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr8_+_42873548 | 0.30 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr3_-_49466686 | 0.29 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr6_-_116833500 | 0.29 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr5_-_111093167 | 0.28 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr22_+_31003190 | 0.28 |
ENST00000407817.3
|
TCN2
|
transcobalamin II |
| chr5_+_67586465 | 0.27 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_+_31002779 | 0.27 |
ENST00000215838.3
|
TCN2
|
transcobalamin II |
| chr19_+_41305085 | 0.26 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr6_+_31707725 | 0.26 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr2_+_234526272 | 0.26 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_+_180319918 | 0.24 |
ENST00000296015.4
ENST00000491380.1 ENST00000412756.2 ENST00000382584.4 |
TTC14
|
tetratricopeptide repeat domain 14 |
| chr3_-_196911002 | 0.24 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr12_+_80838126 | 0.23 |
ENST00000266688.5
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q |
| chr4_+_108815402 | 0.22 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr14_+_95027772 | 0.22 |
ENST00000555095.1
ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr17_-_26733604 | 0.21 |
ENST00000584426.1
ENST00000584995.1 |
SLC46A1
|
solute carrier family 46 (folate transporter), member 1 |
| chrX_+_99899180 | 0.20 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr3_+_66271410 | 0.18 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr19_-_11639931 | 0.16 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr7_-_44580861 | 0.16 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr12_-_15865844 | 0.16 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr15_+_58724184 | 0.14 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr3_-_137893721 | 0.14 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr2_-_180129484 | 0.13 |
ENST00000428443.3
|
SESTD1
|
SEC14 and spectrin domains 1 |
| chr5_+_140571902 | 0.13 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr22_+_31003133 | 0.12 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr17_+_48823896 | 0.12 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_+_84630645 | 0.11 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_+_39703112 | 0.10 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr22_-_22337154 | 0.09 |
ENST00000413067.2
ENST00000437929.1 ENST00000456075.1 ENST00000434517.1 ENST00000424393.1 ENST00000449704.1 ENST00000437103.1 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr21_-_47575481 | 0.09 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr2_-_88427568 | 0.09 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr2_-_74555350 | 0.08 |
ENST00000444570.1
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr4_+_39408470 | 0.08 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr1_-_36020531 | 0.08 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr12_-_56615693 | 0.07 |
ENST00000394013.2
ENST00000345093.4 ENST00000551711.1 ENST00000552656.1 |
RNF41
|
ring finger protein 41 |
| chr17_-_49021974 | 0.07 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr4_+_74269956 | 0.06 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr7_-_47578840 | 0.06 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr2_+_234601512 | 0.05 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr11_+_64358722 | 0.05 |
ENST00000336464.7
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12 |
| chr16_+_20462783 | 0.04 |
ENST00000574251.1
ENST00000576361.1 ENST00000417235.2 ENST00000573854.1 ENST00000424070.1 ENST00000536134.1 ENST00000219054.6 ENST00000575690.1 ENST00000571894.1 |
ACSM2A
|
acyl-CoA synthetase medium-chain family member 2A |
| chr12_-_54653313 | 0.04 |
ENST00000550411.1
ENST00000439541.2 |
CBX5
|
chromobox homolog 5 |
| chr3_+_72200408 | 0.04 |
ENST00000473713.1
|
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr8_+_21911054 | 0.04 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr4_-_74486109 | 0.04 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_-_133084580 | 0.03 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr1_+_207262578 | 0.03 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr11_+_2405833 | 0.02 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF1A_HNF1B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 21.1 | GO:1903412 | response to bile acid(GO:1903412) |
| 2.2 | 8.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.2 | 6.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 1.8 | 5.5 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 1.7 | 18.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 1.6 | 21.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.6 | 7.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 1.4 | 4.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 1.3 | 3.9 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.2 | 6.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.2 | 4.8 | GO:0090293 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 1.2 | 15.6 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 1.0 | 4.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 1.0 | 9.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.9 | 2.8 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.7 | 4.6 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.6 | 1.8 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.5 | 5.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.4 | 1.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 1.1 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) sclerotome development(GO:0061056) |
| 0.3 | 1.7 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.3 | 2.4 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 4.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 10.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 8.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 13.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 1.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 1.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.8 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 1.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 8.7 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.8 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 1.4 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.1 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 1.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 2.7 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 6.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 26.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.5 | 8.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.4 | 4.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.7 | 12.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 3.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 1.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.6 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 0.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 4.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 8.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 5.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 9.1 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 19.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 5.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 26.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 7.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 18.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 8.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 4.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.8 | 11.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 2.1 | 6.2 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 1.9 | 5.7 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 1.8 | 21.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.7 | 10.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 1.3 | 3.9 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.2 | 4.8 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 1.1 | 3.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 1.0 | 7.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.9 | 8.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.8 | 4.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 2.8 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.5 | 1.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 1.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.3 | 1.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.3 | 1.7 | GO:0016160 | amylase activity(GO:0016160) |
| 0.3 | 1.9 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.3 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 3.9 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.2 | 8.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.6 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 11.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 2.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 1.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 6.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 14.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 10.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 18.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 0.3 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 2.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 8.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 4.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 10.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 18.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 31.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 4.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 28.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.3 | 21.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.8 | 22.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 8.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 6.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 2.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 3.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 5.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |