Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
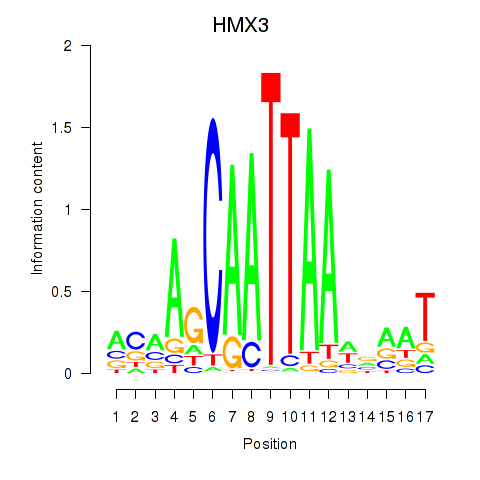
Results for HMX3
Z-value: 1.07
Transcription factors associated with HMX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX3
|
ENSG00000188620.9 | H6 family homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX3 | hg19_v2_chr10_+_124895472_124895508 | 0.57 | 4.3e-01 | Click! |
Activity profile of HMX3 motif
Sorted Z-values of HMX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_149192475 | 1.65 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr17_-_64225508 | 1.58 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_+_186330712 | 1.49 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr1_+_241695670 | 0.83 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_+_241695424 | 0.79 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr5_+_138611798 | 0.76 |
ENST00000502394.1
|
MATR3
|
matrin 3 |
| chr11_+_65266507 | 0.61 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr1_-_86861660 | 0.60 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr8_-_17752912 | 0.60 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr21_+_17909594 | 0.59 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_22517166 | 0.59 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr19_-_13900972 | 0.58 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr7_-_35013217 | 0.58 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr18_+_29171689 | 0.56 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr6_-_52628271 | 0.56 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr8_-_17752996 | 0.56 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr4_-_155511887 | 0.53 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr12_-_10978957 | 0.51 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr9_+_5231413 | 0.50 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr4_-_105416039 | 0.50 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr6_+_26204825 | 0.47 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr12_+_20968608 | 0.46 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr2_-_55237484 | 0.46 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr2_+_200472779 | 0.45 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr8_+_97506033 | 0.45 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr2_-_191115229 | 0.44 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr17_+_1665253 | 0.44 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr15_-_31521567 | 0.44 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr16_+_20775358 | 0.44 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr11_+_327171 | 0.44 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr7_-_7680601 | 0.43 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr4_-_76649546 | 0.43 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_+_26124373 | 0.43 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr7_-_16840820 | 0.43 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr12_-_27167233 | 0.42 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr8_-_93978357 | 0.42 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr15_+_69857515 | 0.40 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr21_+_17791648 | 0.39 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_-_56757329 | 0.39 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr8_-_101571964 | 0.38 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr1_-_149459549 | 0.38 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr14_+_74551650 | 0.38 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr12_+_100867486 | 0.38 |
ENST00000548884.1
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_+_139528952 | 0.38 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr15_+_58702742 | 0.37 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr6_-_132272504 | 0.37 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr16_+_32264040 | 0.36 |
ENST00000398664.3
|
TP53TG3D
|
TP53 target 3D |
| chr1_-_150978953 | 0.35 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr16_+_53412368 | 0.35 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr19_+_52772821 | 0.35 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr11_+_115498761 | 0.35 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr16_+_20775024 | 0.35 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr12_+_147052 | 0.35 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr10_-_101945771 | 0.35 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr21_-_39705323 | 0.35 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr1_-_108231101 | 0.34 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr8_-_27695552 | 0.34 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr7_-_130066571 | 0.34 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr19_-_16008880 | 0.34 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr4_-_104021009 | 0.33 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr2_-_134326009 | 0.33 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr19_+_42212526 | 0.33 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr4_+_25915896 | 0.32 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr3_+_195413160 | 0.32 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr1_-_220263096 | 0.32 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr4_-_76944621 | 0.32 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr16_+_10479906 | 0.32 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr2_+_128293323 | 0.31 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr1_-_197036364 | 0.31 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr5_+_95066823 | 0.31 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr16_-_46655538 | 0.30 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr3_+_39424828 | 0.30 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr6_-_151773232 | 0.30 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr1_+_109289279 | 0.30 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr17_-_4938712 | 0.29 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr7_-_111032971 | 0.29 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_+_28299014 | 0.29 |
ENST00000538586.1
ENST00000536154.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_123717711 | 0.29 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr2_+_108994466 | 0.29 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr1_-_63782888 | 0.29 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr13_+_48611665 | 0.28 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr5_+_102594403 | 0.28 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chrX_+_38420623 | 0.28 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr15_+_40674920 | 0.28 |
ENST00000416151.2
ENST00000249776.8 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr8_-_95487331 | 0.28 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr11_+_827248 | 0.28 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr14_-_92247032 | 0.27 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr12_-_91573132 | 0.27 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr8_-_101571933 | 0.27 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr12_-_89919965 | 0.27 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr19_+_49199209 | 0.27 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr8_+_109455830 | 0.27 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr5_+_156696362 | 0.27 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_-_65426174 | 0.27 |
ENST00000204549.4
|
PDCD7
|
programmed cell death 7 |
| chr13_+_110958124 | 0.27 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr2_+_158114051 | 0.27 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr2_+_242289502 | 0.26 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr17_-_46671323 | 0.26 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr10_-_75168071 | 0.26 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr6_+_29624758 | 0.26 |
ENST00000376917.3
ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr1_+_53480598 | 0.26 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr22_+_39795746 | 0.26 |
ENST00000216160.6
ENST00000331454.3 |
TAB1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr8_-_93978333 | 0.26 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr7_-_84569561 | 0.26 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr19_+_52772832 | 0.26 |
ENST00000593703.1
ENST00000601711.1 ENST00000599581.1 |
ZNF766
|
zinc finger protein 766 |
| chr2_+_128177458 | 0.26 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr17_+_68071389 | 0.26 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_+_11333245 | 0.26 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr1_-_21113105 | 0.26 |
ENST00000375000.1
ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr8_+_38758737 | 0.25 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr21_-_35773370 | 0.25 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr14_+_64680854 | 0.25 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr3_-_87325612 | 0.25 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr14_-_74025625 | 0.25 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr7_-_92146729 | 0.25 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr5_+_112849373 | 0.25 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chrX_-_107682702 | 0.24 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr9_+_5890802 | 0.24 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr22_+_23487513 | 0.24 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr5_-_74162605 | 0.24 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr4_-_104020968 | 0.24 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr2_-_152590982 | 0.24 |
ENST00000409198.1
ENST00000397345.3 ENST00000427231.2 |
NEB
|
nebulin |
| chr12_-_89920030 | 0.24 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr7_-_32529973 | 0.24 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr4_+_109571740 | 0.24 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr18_+_2571510 | 0.24 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr14_-_21566731 | 0.24 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr5_+_82373379 | 0.24 |
ENST00000396027.4
ENST00000511817.1 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr3_-_151176497 | 0.24 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr6_-_26056695 | 0.23 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chrX_+_38420783 | 0.23 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr19_-_58204128 | 0.23 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr6_-_52668605 | 0.23 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr15_-_55700457 | 0.23 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr2_+_122513109 | 0.23 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr7_+_139529040 | 0.23 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr6_+_25652432 | 0.23 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr3_-_165555200 | 0.23 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr5_+_82373317 | 0.23 |
ENST00000282268.3
ENST00000338635.6 |
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr2_+_44001172 | 0.23 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr7_+_77325738 | 0.23 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr3_+_121613265 | 0.23 |
ENST00000295605.2
|
SLC15A2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chrX_-_15511438 | 0.23 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr3_-_114343039 | 0.23 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_220445757 | 0.23 |
ENST00000358951.2
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chrX_+_72783026 | 0.23 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr15_+_67835517 | 0.23 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr17_+_68100989 | 0.23 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr8_-_124749609 | 0.22 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr12_-_15374343 | 0.22 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr4_-_110723194 | 0.22 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr10_+_116697946 | 0.22 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr4_-_110723134 | 0.22 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr4_-_110624564 | 0.22 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr14_+_57857262 | 0.22 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr5_-_111754948 | 0.21 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr19_+_58144529 | 0.21 |
ENST00000347302.3
ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211
|
zinc finger protein 211 |
| chr1_+_100598691 | 0.21 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr1_+_45205591 | 0.21 |
ENST00000455186.1
|
KIF2C
|
kinesin family member 2C |
| chr12_+_11081828 | 0.21 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr17_+_53016208 | 0.21 |
ENST00000574318.1
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr11_+_125496619 | 0.21 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr1_-_243326612 | 0.20 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr5_+_175288631 | 0.20 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr12_+_100867694 | 0.20 |
ENST00000392986.3
ENST00000549996.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_-_47465666 | 0.20 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr3_+_124223586 | 0.20 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr4_+_129731074 | 0.20 |
ENST00000512960.1
ENST00000503785.1 ENST00000514740.1 |
PHF17
|
jade family PHD finger 1 |
| chr7_-_16921601 | 0.20 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr1_+_45212051 | 0.20 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr5_+_42756903 | 0.20 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr15_-_55700522 | 0.20 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr14_-_90421028 | 0.20 |
ENST00000267544.9
ENST00000316738.7 ENST00000538485.2 ENST00000556609.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr15_+_59903975 | 0.20 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr14_-_90420862 | 0.20 |
ENST00000556005.1
ENST00000555872.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr7_+_91570165 | 0.20 |
ENST00000356239.3
ENST00000359028.2 ENST00000358100.2 |
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr12_+_107712173 | 0.20 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr6_-_26285737 | 0.20 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr8_+_101349823 | 0.20 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr2_+_219472637 | 0.20 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr1_+_101361626 | 0.20 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr18_-_55253989 | 0.20 |
ENST00000262093.5
|
FECH
|
ferrochelatase |
| chr4_+_95128748 | 0.20 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_-_111649015 | 0.19 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr13_-_79233314 | 0.19 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr1_+_201979645 | 0.19 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr5_+_162930114 | 0.19 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr2_+_90458201 | 0.19 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr1_+_109102652 | 0.19 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr15_-_35838348 | 0.19 |
ENST00000561411.1
ENST00000256538.4 ENST00000440392.2 |
DPH6
|
diphthamine biosynthesis 6 |
| chr6_+_12290586 | 0.19 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr12_+_27863706 | 0.19 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr18_-_46895066 | 0.19 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr20_-_8000426 | 0.19 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr12_-_52604607 | 0.19 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr4_-_48782259 | 0.19 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr1_+_81106951 | 0.19 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr2_+_108905325 | 0.19 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr3_-_112738490 | 0.18 |
ENST00000393857.2
|
C3orf17
|
chromosome 3 open reading frame 17 |
| chr3_-_148939835 | 0.18 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:0090293 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 1.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.4 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 1.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.3 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.2 | GO:1904346 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.3 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.2 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.3 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.3 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0060516 | primary prostatic bud elongation(GO:0060516) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.3 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 1.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0071262 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:1904646 | response to beta-amyloid(GO:1904645) cellular response to beta-amyloid(GO:1904646) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.0 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.3 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 1.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0070409 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.9 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.0 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.0 | GO:0009804 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.5 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0098552 | external side of plasma membrane(GO:0009897) side of membrane(GO:0098552) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.7 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.6 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 0.9 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.2 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.1 | 0.2 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.2 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.4 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |