Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for HMX2
Z-value: 1.14
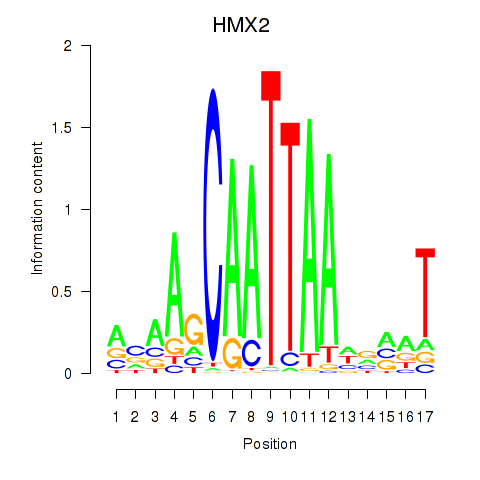
Transcription factors associated with HMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX2
|
ENSG00000188816.3 | H6 family homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX2 | hg19_v2_chr10_+_124907638_124907660 | -0.27 | 7.3e-01 | Click! |
Activity profile of HMX2 motif
Sorted Z-values of HMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_327537 | 1.41 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_+_5710919 | 1.26 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr22_+_50981079 | 1.19 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr15_-_60695071 | 0.84 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr19_-_3557570 | 0.62 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr6_-_27858570 | 0.60 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr10_-_126849626 | 0.59 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr15_+_101417919 | 0.52 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr1_-_202858227 | 0.51 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr14_+_101295638 | 0.51 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr14_-_101295407 | 0.50 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr18_+_68002675 | 0.50 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr6_-_26124138 | 0.49 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr19_+_9361606 | 0.49 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr9_-_14322319 | 0.48 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr2_+_45168875 | 0.48 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr1_+_209878182 | 0.47 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr20_+_42136308 | 0.46 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr10_-_22292675 | 0.42 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr1_-_110933611 | 0.42 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr11_+_110225855 | 0.41 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr4_-_120243545 | 0.40 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr21_-_39705286 | 0.39 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chrX_-_1331527 | 0.38 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr17_-_29641104 | 0.37 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr12_-_94673956 | 0.35 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr1_+_239882842 | 0.35 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr8_-_93978309 | 0.34 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr13_+_49551020 | 0.33 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr15_+_41056218 | 0.33 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr6_-_160209471 | 0.32 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr19_-_51568324 | 0.32 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr3_+_38537960 | 0.32 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr9_+_26746951 | 0.32 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr18_+_76829441 | 0.31 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr4_-_159080806 | 0.30 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr11_+_60681346 | 0.30 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr6_+_77484663 | 0.30 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr14_+_55595762 | 0.29 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr14_-_50864006 | 0.27 |
ENST00000216378.2
|
CDKL1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chrX_-_101186981 | 0.27 |
ENST00000458570.1
|
ZMAT1
|
zinc finger, matrin-type 1 |
| chr8_+_32579271 | 0.27 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr1_-_110933663 | 0.27 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr11_-_74442430 | 0.26 |
ENST00000376332.3
|
CHRDL2
|
chordin-like 2 |
| chr11_-_64684672 | 0.26 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr12_-_49463753 | 0.26 |
ENST00000301068.6
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr22_+_42095497 | 0.26 |
ENST00000401548.3
ENST00000540833.1 ENST00000400107.1 ENST00000300398.4 |
MEI1
|
meiosis inhibitor 1 |
| chr4_+_156775910 | 0.25 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr17_-_15469590 | 0.25 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr9_+_130159433 | 0.25 |
ENST00000451404.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr12_-_112123524 | 0.25 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr20_-_47804894 | 0.24 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr3_+_23847394 | 0.24 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr12_+_64798095 | 0.23 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr4_-_90759440 | 0.23 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_94277017 | 0.23 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chrX_+_36254051 | 0.22 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr2_+_211342400 | 0.22 |
ENST00000417946.1
ENST00000518043.1 ENST00000523702.1 |
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr1_+_182584314 | 0.22 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr4_+_70894130 | 0.22 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr8_+_32579321 | 0.21 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr1_-_11918988 | 0.21 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr3_+_145782358 | 0.21 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr10_-_49813090 | 0.21 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr17_-_29641084 | 0.21 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr11_-_128894053 | 0.20 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_+_185303962 | 0.20 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr10_-_95241951 | 0.20 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_-_187009646 | 0.20 |
ENST00000296280.6
ENST00000392470.2 ENST00000169293.6 ENST00000439271.1 ENST00000392472.2 ENST00000392475.2 |
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr11_-_18211042 | 0.20 |
ENST00000527671.1
|
RP11-113D6.6
|
Uncharacterized protein |
| chr10_+_12238171 | 0.20 |
ENST00000378900.2
ENST00000442050.1 |
CDC123
|
cell division cycle 123 |
| chrX_+_1710484 | 0.20 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr5_-_43397184 | 0.20 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr2_-_145277569 | 0.19 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_122985067 | 0.19 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr6_-_26199499 | 0.19 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr12_-_72057571 | 0.19 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr19_+_40877583 | 0.19 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr15_+_54305101 | 0.19 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr11_+_64685026 | 0.18 |
ENST00000526559.1
|
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr12_-_49463620 | 0.18 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr19_+_52772832 | 0.18 |
ENST00000593703.1
ENST00000601711.1 ENST00000599581.1 |
ZNF766
|
zinc finger protein 766 |
| chr4_+_166248775 | 0.18 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chrX_+_134478706 | 0.17 |
ENST00000370761.3
ENST00000339249.4 ENST00000370760.3 |
ZNF449
|
zinc finger protein 449 |
| chr5_+_61602236 | 0.17 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr20_+_5986756 | 0.17 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr3_-_194188956 | 0.17 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr1_-_109399682 | 0.16 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr12_+_128399965 | 0.16 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr8_+_31496809 | 0.16 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chrX_+_36053908 | 0.16 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr5_-_58295712 | 0.15 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_-_151344172 | 0.15 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr19_-_52674896 | 0.15 |
ENST00000322146.8
ENST00000597065.1 |
ZNF836
|
zinc finger protein 836 |
| chr4_+_83956312 | 0.15 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr4_-_69817481 | 0.15 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr3_-_79816965 | 0.15 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_3772209 | 0.15 |
ENST00000555978.1
ENST00000555633.1 |
RAX2
|
retina and anterior neural fold homeobox 2 |
| chr6_+_117198400 | 0.15 |
ENST00000332958.2
|
RFX6
|
regulatory factor X, 6 |
| chr3_+_122785895 | 0.15 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr6_+_151561085 | 0.14 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr19_+_18668616 | 0.14 |
ENST00000600372.1
|
KXD1
|
KxDL motif containing 1 |
| chr14_-_73997901 | 0.14 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr8_+_77318769 | 0.14 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr13_-_103426112 | 0.14 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr1_+_40810516 | 0.14 |
ENST00000435168.2
|
SMAP2
|
small ArfGAP2 |
| chr10_+_81462983 | 0.14 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr1_+_33546714 | 0.13 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr11_-_8857248 | 0.13 |
ENST00000534248.1
ENST00000530959.1 ENST00000531578.1 ENST00000533225.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr2_+_24272543 | 0.13 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr2_-_211342292 | 0.13 |
ENST00000448951.1
|
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr3_+_23851928 | 0.13 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr12_-_13248598 | 0.13 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr9_-_111882195 | 0.13 |
ENST00000374586.3
|
TMEM245
|
transmembrane protein 245 |
| chr6_-_53481847 | 0.13 |
ENST00000503985.1
|
RP1-27K12.2
|
RP1-27K12.2 |
| chr14_+_65381079 | 0.13 |
ENST00000549115.1
ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1
FNTB
CHURC1-FNTB
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr14_-_20801427 | 0.12 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr6_-_51952367 | 0.12 |
ENST00000340994.4
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr2_+_172309634 | 0.12 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr2_-_171571077 | 0.12 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr12_+_128399917 | 0.12 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr3_-_187009798 | 0.12 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr8_+_32579341 | 0.12 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr3_+_173116225 | 0.12 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr4_-_168155169 | 0.12 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_-_51952418 | 0.12 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr17_-_73663245 | 0.12 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr9_+_130159409 | 0.12 |
ENST00000373371.3
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr21_+_44073860 | 0.11 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr1_+_144151520 | 0.11 |
ENST00000369372.4
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr2_-_130031335 | 0.11 |
ENST00000375987.3
|
AC079586.1
|
AC079586.1 |
| chr7_-_20256965 | 0.11 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr3_+_16216137 | 0.11 |
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr7_-_87856280 | 0.11 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr20_+_3776371 | 0.11 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr4_-_39034542 | 0.11 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chrX_-_153979315 | 0.11 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr14_+_92789498 | 0.11 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr3_-_112013071 | 0.11 |
ENST00000487372.1
ENST00000486574.1 ENST00000305815.5 |
SLC9C1
|
solute carrier family 9, subfamily C (Na+-transporting carboxylic acid decarboxylase), member 1 |
| chr1_-_217804377 | 0.11 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr13_-_103426081 | 0.11 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr2_+_109237717 | 0.10 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr19_-_51340469 | 0.10 |
ENST00000326856.4
|
KLK15
|
kallikrein-related peptidase 15 |
| chr10_-_95242044 | 0.10 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr11_+_105480793 | 0.10 |
ENST00000282499.5
ENST00000393127.2 ENST00000527669.1 |
GRIA4
|
glutamate receptor, ionotropic, AMPA 4 |
| chr1_-_21377447 | 0.10 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr17_+_48823896 | 0.10 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr2_-_111334678 | 0.10 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr1_+_70820451 | 0.10 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr14_+_64565442 | 0.10 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr9_-_116102530 | 0.09 |
ENST00000374195.3
ENST00000341761.4 |
WDR31
|
WD repeat domain 31 |
| chr19_+_18668572 | 0.09 |
ENST00000540691.1
ENST00000539106.1 ENST00000222307.4 |
KXD1
|
KxDL motif containing 1 |
| chr19_+_12175504 | 0.09 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr3_+_179280668 | 0.09 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr11_-_118972575 | 0.09 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr19_-_44384291 | 0.09 |
ENST00000324394.6
|
ZNF404
|
zinc finger protein 404 |
| chr3_+_101818088 | 0.09 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr2_-_20251744 | 0.09 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr2_+_27071045 | 0.09 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr21_+_44073916 | 0.09 |
ENST00000349112.3
ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr19_-_36001113 | 0.09 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr14_-_59950724 | 0.09 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr3_+_186560462 | 0.09 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr14_+_32798462 | 0.09 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr18_+_76829385 | 0.08 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr8_-_93978346 | 0.08 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr15_+_55700741 | 0.08 |
ENST00000569691.1
|
C15orf65
|
chromosome 15 open reading frame 65 |
| chr15_-_40074996 | 0.08 |
ENST00000350221.3
|
FSIP1
|
fibrous sheath interacting protein 1 |
| chr15_-_50558223 | 0.08 |
ENST00000267845.3
|
HDC
|
histidine decarboxylase |
| chr17_-_76824975 | 0.08 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr5_+_140227048 | 0.08 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr2_+_234826016 | 0.08 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr6_-_106773491 | 0.08 |
ENST00000360666.4
|
ATG5
|
autophagy related 5 |
| chr5_+_154393260 | 0.08 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr7_-_87856303 | 0.08 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr7_+_2393714 | 0.08 |
ENST00000431643.1
|
EIF3B
|
eukaryotic translation initiation factor 3, subunit B |
| chr18_+_10526008 | 0.08 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr2_+_27071292 | 0.08 |
ENST00000431402.1
ENST00000434719.1 |
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr10_-_27529486 | 0.08 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr14_+_31091511 | 0.08 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chr14_+_55595960 | 0.07 |
ENST00000554715.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr14_+_50291993 | 0.07 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr8_-_93978333 | 0.07 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_125811162 | 0.07 |
ENST00000299308.3
|
TMEM132B
|
transmembrane protein 132B |
| chr6_-_46138676 | 0.07 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr1_-_205290865 | 0.07 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr1_+_196946664 | 0.07 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr3_+_186560476 | 0.07 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr10_-_126849588 | 0.07 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr7_+_39125365 | 0.07 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr2_+_108994633 | 0.07 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr10_-_115613828 | 0.07 |
ENST00000361384.2
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr5_+_167913450 | 0.06 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr1_+_196946680 | 0.06 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr10_-_22292613 | 0.06 |
ENST00000376980.3
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr14_+_73706308 | 0.06 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr6_-_116833500 | 0.06 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr2_-_169104651 | 0.06 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr3_+_156799587 | 0.06 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr19_-_14889349 | 0.06 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 0.8 | GO:0052362 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.2 | 0.5 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.4 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.3 | GO:0044417 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.2 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.2 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 1.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.2 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 1.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.0 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.3 | GO:0045141 | meiotic telomere clustering(GO:0045141) |
| 0.0 | 0.2 | GO:0035562 | protein desumoylation(GO:0016926) negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.5 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.7 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |