Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
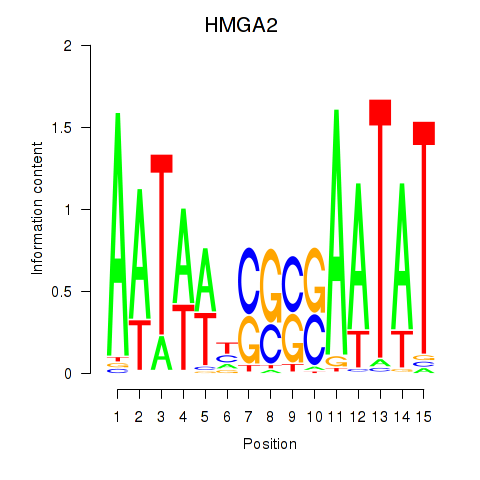
Results for HMGA2
Z-value: 1.77
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | high mobility group AT-hook 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA2 | hg19_v2_chr12_+_66218598_66218678 | 0.65 | 3.5e-01 | Click! |
Activity profile of HMGA2 motif
Sorted Z-values of HMGA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_64014588 | 2.32 |
ENST00000371086.2
ENST00000340052.3 |
DLEU2L
|
deleted in lymphocytic leukemia 2-like |
| chr16_+_53483983 | 1.69 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr19_-_36297632 | 1.69 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr6_-_107230334 | 1.66 |
ENST00000607090.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr1_+_196621002 | 1.64 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr2_+_13677795 | 1.47 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr6_-_32908792 | 1.35 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr13_+_50656307 | 1.31 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr15_+_58702742 | 1.25 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr16_+_66442411 | 1.21 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr1_+_196621156 | 1.08 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr6_-_32908765 | 0.98 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chrX_+_117480036 | 0.98 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr3_+_186501336 | 0.93 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr1_+_179263308 | 0.89 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr18_-_14132422 | 0.79 |
ENST00000589498.1
ENST00000590202.1 |
ZNF519
|
zinc finger protein 519 |
| chr10_+_96305535 | 0.78 |
ENST00000419900.1
ENST00000348459.5 ENST00000394045.1 ENST00000394044.1 ENST00000394036.1 |
HELLS
|
helicase, lymphoid-specific |
| chr13_+_51483814 | 0.75 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr1_+_227751231 | 0.73 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr9_-_115480303 | 0.72 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr14_-_94759595 | 0.71 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr1_+_28836561 | 0.69 |
ENST00000419074.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr1_-_226374373 | 0.69 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr1_+_104104379 | 0.66 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr3_-_123680246 | 0.62 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr6_-_136571400 | 0.61 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chrX_-_135962876 | 0.61 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr9_+_2157655 | 0.60 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_116892641 | 0.60 |
ENST00000487832.2
ENST00000518117.1 |
RWDD1
|
RWD domain containing 1 |
| chr6_-_160209471 | 0.59 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr14_-_94759361 | 0.59 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759408 | 0.59 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr20_+_48884002 | 0.57 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr7_-_102985035 | 0.56 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr17_-_76220740 | 0.55 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr2_+_177134201 | 0.54 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr10_+_86184676 | 0.51 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr5_-_102455801 | 0.50 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr1_+_2487631 | 0.50 |
ENST00000409119.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr12_-_122750957 | 0.49 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr14_-_65409502 | 0.48 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr3_+_106959530 | 0.48 |
ENST00000466734.1
ENST00000463143.1 ENST00000490441.1 |
LINC00883
|
long intergenic non-protein coding RNA 883 |
| chr9_+_42704004 | 0.47 |
ENST00000457288.1
|
CBWD7
|
COBW domain containing 7 |
| chr6_+_111580508 | 0.47 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr1_+_55464600 | 0.47 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr7_-_102985288 | 0.45 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr15_+_82722225 | 0.42 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr19_-_38146289 | 0.42 |
ENST00000392144.1
ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30
|
ZFP30 zinc finger protein |
| chr2_-_216257849 | 0.41 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr14_-_65409438 | 0.40 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr15_+_83098710 | 0.38 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chr1_-_223988426 | 0.38 |
ENST00000391879.2
|
TP53BP2
|
tumor protein p53 binding protein, 2 |
| chr10_-_101673782 | 0.37 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr2_+_170683979 | 0.37 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr2_+_177134134 | 0.36 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr14_+_45553296 | 0.35 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr6_-_127664736 | 0.35 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr2_+_172543967 | 0.35 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr15_-_32747835 | 0.32 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chrX_-_135962923 | 0.32 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr14_+_99947715 | 0.32 |
ENST00000389879.5
ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK
|
cyclin K |
| chr14_+_88852059 | 0.31 |
ENST00000045347.7
|
SPATA7
|
spermatogenesis associated 7 |
| chr15_+_84904525 | 0.31 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr18_+_11851383 | 0.30 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr2_+_120436733 | 0.29 |
ENST00000401466.1
ENST00000424086.1 ENST00000272521.6 |
TMEM177
|
transmembrane protein 177 |
| chr15_+_28623784 | 0.29 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr6_-_127664683 | 0.28 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr2_+_172544011 | 0.27 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr15_-_70994612 | 0.27 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr2_+_172543919 | 0.26 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_+_65996599 | 0.25 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr19_+_41882466 | 0.22 |
ENST00000436170.2
|
TMEM91
|
transmembrane protein 91 |
| chr4_+_25378912 | 0.21 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr15_+_43885252 | 0.21 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr9_+_104296122 | 0.21 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr12_-_8765446 | 0.21 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chrX_-_77150985 | 0.20 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr3_+_106959552 | 0.20 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883 |
| chr6_+_106535455 | 0.19 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr10_-_36813162 | 0.19 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr2_+_172544182 | 0.18 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr17_+_73230799 | 0.17 |
ENST00000579838.1
|
NUP85
|
nucleoporin 85kDa |
| chr6_+_88106840 | 0.17 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr19_-_1650666 | 0.15 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr3_+_193853927 | 0.15 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr19_+_44455368 | 0.15 |
ENST00000591168.1
ENST00000587682.1 ENST00000251269.5 |
ZNF221
|
zinc finger protein 221 |
| chr15_+_43985084 | 0.15 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr17_-_295730 | 0.14 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr21_+_38792602 | 0.14 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr15_-_74659978 | 0.10 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr6_+_123110302 | 0.10 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr12_+_59194154 | 0.09 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr11_-_130786400 | 0.09 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr12_+_113587558 | 0.09 |
ENST00000335621.6
|
CCDC42B
|
coiled-coil domain containing 42B |
| chr19_+_11457175 | 0.08 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chr2_+_120436760 | 0.08 |
ENST00000445518.1
ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr12_+_21207503 | 0.07 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr19_-_43099070 | 0.05 |
ENST00000244336.5
|
CEACAM8
|
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr2_-_114461655 | 0.04 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chrX_-_7895755 | 0.03 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr10_-_62332357 | 0.03 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_+_170683942 | 0.02 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr5_-_135290705 | 0.02 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr12_-_21757774 | 0.01 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:2001190 | peptide antigen assembly with MHC class II protein complex(GO:0002503) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.4 | 1.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 0.8 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.2 | 1.7 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.0 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 0.8 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.2 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0044417 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.9 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.4 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 1.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 1.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.2 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 0.2 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) regulation of timing of neuron differentiation(GO:0060164) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 1.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 2.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 0.9 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 0.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.6 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 1.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 2.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 2.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 1.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 3.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |