Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
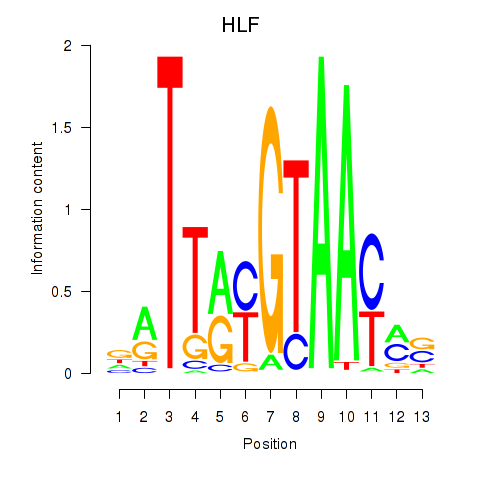
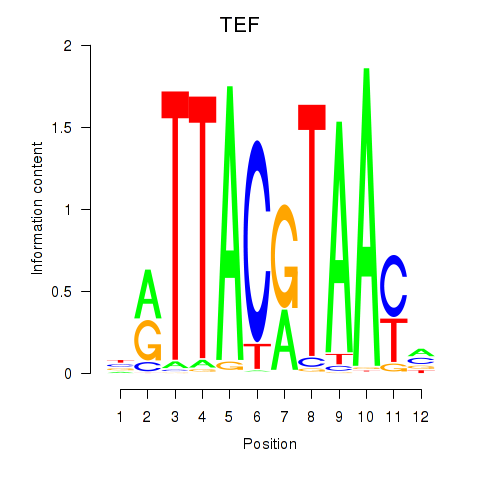
Results for HLF_TEF
Z-value: 1.25
Transcription factors associated with HLF_TEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HLF
|
ENSG00000108924.9 | HLF transcription factor, PAR bZIP family member |
|
TEF
|
ENSG00000167074.10 | TEF transcription factor, PAR bZIP family member |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLF | hg19_v2_chr17_+_53343577_53343588 | 1.00 | 3.4e-03 | Click! |
| TEF | hg19_v2_chr22_+_41763274_41763337 | 0.92 | 7.7e-02 | Click! |
Activity profile of HLF_TEF motif
Sorted Z-values of HLF_TEF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_158390282 | 1.79 |
ENST00000264265.3
|
LXN
|
latexin |
| chr1_+_154377669 | 1.30 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr20_+_4666882 | 1.06 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr3_+_186330712 | 1.00 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr17_+_70036164 | 0.97 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr1_+_7844312 | 0.95 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr8_-_27469383 | 0.85 |
ENST00000519742.1
|
CLU
|
clusterin |
| chr1_-_226496772 | 0.81 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr20_+_4667094 | 0.77 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr8_-_27468842 | 0.76 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr1_+_212738676 | 0.76 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr16_-_53537105 | 0.75 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr13_-_46626820 | 0.72 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr14_+_75894391 | 0.69 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr8_-_27468717 | 0.68 |
ENST00000520796.1
ENST00000520491.1 |
CLU
|
clusterin |
| chr8_-_27469196 | 0.68 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr8_-_27468945 | 0.66 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr5_+_95997918 | 0.60 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr1_+_229440129 | 0.57 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr16_-_54962415 | 0.56 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr6_-_86353510 | 0.55 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr13_-_46626847 | 0.53 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr5_+_95998070 | 0.52 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr7_+_90032667 | 0.48 |
ENST00000496677.1
ENST00000287916.4 ENST00000535571.1 ENST00000394604.1 ENST00000394605.2 |
CLDN12
|
claudin 12 |
| chr16_-_54962704 | 0.48 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_+_65613513 | 0.47 |
ENST00000395334.2
|
AK4
|
adenylate kinase 4 |
| chr1_-_114429997 | 0.46 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr5_+_95997769 | 0.46 |
ENST00000338252.3
ENST00000508830.1 |
CAST
|
calpastatin |
| chr5_+_95997885 | 0.44 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr7_+_23145884 | 0.42 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr14_-_23284703 | 0.41 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_-_141401951 | 0.41 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr19_-_2042065 | 0.41 |
ENST00000591588.1
ENST00000591142.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr3_+_101504200 | 0.41 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr4_+_157997273 | 0.39 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr18_+_32621324 | 0.37 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr5_+_172332220 | 0.37 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr14_-_23285069 | 0.37 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr10_+_94050913 | 0.37 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr2_+_183989157 | 0.36 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr14_-_23285011 | 0.35 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_-_58200398 | 0.34 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr9_+_71820057 | 0.34 |
ENST00000539225.1
|
TJP2
|
tight junction protein 2 |
| chr1_+_196621156 | 0.33 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr17_-_19651598 | 0.33 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr18_+_3262954 | 0.33 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr13_-_45010939 | 0.33 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr8_+_19536083 | 0.32 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr2_-_167232484 | 0.31 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr9_+_71819927 | 0.30 |
ENST00000535702.1
|
TJP2
|
tight junction protein 2 |
| chr9_-_123639445 | 0.30 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr1_+_90308981 | 0.30 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr18_-_55288973 | 0.30 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr8_+_107738240 | 0.30 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr9_-_34665983 | 0.29 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr9_-_123639304 | 0.27 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr9_-_88356789 | 0.27 |
ENST00000357081.3
ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr11_+_105948216 | 0.26 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr16_+_70680439 | 0.26 |
ENST00000288098.2
|
IL34
|
interleukin 34 |
| chr4_-_153274078 | 0.26 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr15_+_49913201 | 0.25 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr17_-_26941179 | 0.25 |
ENST00000301037.5
ENST00000530121.1 ENST00000525510.1 ENST00000577790.1 ENST00000531839.1 ENST00000534850.1 |
SGK494
RP11-192H23.4
|
uncharacterized serine/threonine-protein kinase SgK494 Uncharacterized protein |
| chr15_-_83621435 | 0.24 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr9_-_128246769 | 0.24 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr21_+_35107346 | 0.24 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr1_+_45212051 | 0.24 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr7_-_15601595 | 0.23 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr7_-_37026108 | 0.23 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr12_-_122017542 | 0.23 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr20_-_35329063 | 0.23 |
ENST00000422536.1
|
NDRG3
|
NDRG family member 3 |
| chr11_-_33795893 | 0.23 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr4_+_144257915 | 0.23 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr10_-_82049424 | 0.22 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr20_-_49547731 | 0.22 |
ENST00000396029.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr17_-_19651654 | 0.22 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr20_-_49547910 | 0.22 |
ENST00000396032.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr2_-_225362533 | 0.21 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr17_-_41984835 | 0.21 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr2_-_239197201 | 0.21 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr9_-_130712995 | 0.21 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr17_-_36956155 | 0.20 |
ENST00000269554.3
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr1_-_101491319 | 0.20 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr3_+_88188254 | 0.20 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr17_-_73179046 | 0.20 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr8_+_110552831 | 0.19 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr16_+_3508063 | 0.19 |
ENST00000576787.1
ENST00000572942.1 ENST00000576916.1 ENST00000575076.1 ENST00000572131.1 |
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr2_+_46769798 | 0.19 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr17_-_41623075 | 0.19 |
ENST00000545089.1
|
ETV4
|
ets variant 4 |
| chr19_-_35992780 | 0.19 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr17_-_19651668 | 0.18 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr18_+_21032781 | 0.18 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr19_+_50922187 | 0.18 |
ENST00000595883.1
ENST00000597855.1 ENST00000596074.1 ENST00000439922.2 ENST00000594685.1 ENST00000270632.7 |
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr9_-_123639600 | 0.17 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr16_+_3507985 | 0.17 |
ENST00000421765.3
ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr17_-_41623259 | 0.17 |
ENST00000538265.1
ENST00000591713.1 |
ETV4
|
ets variant 4 |
| chr2_+_103378472 | 0.17 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr4_+_48807155 | 0.17 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr4_+_96012614 | 0.17 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr3_+_148447887 | 0.17 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr12_+_52430894 | 0.17 |
ENST00000546842.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr17_+_39975455 | 0.16 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr4_+_110354928 | 0.16 |
ENST00000504968.2
ENST00000399100.2 ENST00000265175.5 |
SEC24B
|
SEC24 family member B |
| chr1_-_167883327 | 0.16 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr2_+_191208791 | 0.16 |
ENST00000423767.1
ENST00000451089.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_+_45212074 | 0.16 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr10_-_65028938 | 0.16 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr17_-_73178599 | 0.16 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr10_-_65028817 | 0.16 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr8_+_90914073 | 0.15 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr7_+_73868439 | 0.15 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr4_+_41937131 | 0.15 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr11_-_77185094 | 0.15 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr19_+_1450112 | 0.15 |
ENST00000590469.1
ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2
|
adenomatosis polyposis coli 2 |
| chr2_-_214014959 | 0.15 |
ENST00000442445.1
ENST00000457361.1 ENST00000342002.2 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr17_+_61851504 | 0.15 |
ENST00000359353.5
ENST00000389924.2 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr14_+_93260569 | 0.14 |
ENST00000163416.2
|
GOLGA5
|
golgin A5 |
| chr7_-_45026200 | 0.14 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr1_-_114430169 | 0.14 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr5_-_43557791 | 0.14 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr19_-_42947121 | 0.14 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr14_+_60716159 | 0.14 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr17_+_76037081 | 0.14 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr8_+_110552337 | 0.14 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr6_-_64029879 | 0.13 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr21_+_27011584 | 0.13 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr4_+_144258021 | 0.13 |
ENST00000262994.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr17_+_61851157 | 0.13 |
ENST00000578681.1
ENST00000583590.1 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr11_-_105948129 | 0.13 |
ENST00000526793.1
|
KBTBD3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr8_+_42396936 | 0.13 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr21_-_30365136 | 0.12 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr14_+_93260642 | 0.12 |
ENST00000355976.2
|
GOLGA5
|
golgin A5 |
| chr2_+_191208601 | 0.12 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chrX_+_9431324 | 0.12 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr9_-_88356733 | 0.12 |
ENST00000376083.3
|
AGTPBP1
|
ATP/GTP binding protein 1 |
| chr17_-_61850894 | 0.12 |
ENST00000403162.3
ENST00000582252.1 ENST00000225726.5 |
CCDC47
|
coiled-coil domain containing 47 |
| chr10_-_91295304 | 0.11 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr4_-_65275162 | 0.11 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr8_-_102216925 | 0.11 |
ENST00000517844.1
|
ZNF706
|
zinc finger protein 706 |
| chr4_+_144258288 | 0.11 |
ENST00000514639.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr6_-_74231303 | 0.11 |
ENST00000309268.6
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr2_-_214015111 | 0.11 |
ENST00000433134.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr12_+_48592134 | 0.11 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr4_-_74088800 | 0.11 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr14_-_21489271 | 0.11 |
ENST00000553593.1
|
NDRG2
|
NDRG family member 2 |
| chr3_+_62304648 | 0.10 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr12_-_113574028 | 0.10 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr17_-_34329084 | 0.10 |
ENST00000354059.4
ENST00000536149.1 |
CCL15
CCL14
|
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr12_+_57624085 | 0.10 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_-_98280383 | 0.10 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr7_-_83824449 | 0.09 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr16_-_70323422 | 0.09 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr16_+_56965960 | 0.09 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr19_+_35330037 | 0.09 |
ENST00000601776.1
|
CTC-523E23.5
|
CTC-523E23.5 |
| chr4_-_21950356 | 0.09 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr12_-_120966943 | 0.09 |
ENST00000552443.1
ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5
|
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr16_-_54962625 | 0.09 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr14_+_60715928 | 0.09 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr2_+_183989083 | 0.09 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr14_-_67826538 | 0.08 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr4_-_96470350 | 0.08 |
ENST00000504962.1
ENST00000453304.1 ENST00000506749.1 |
UNC5C
|
unc-5 homolog C (C. elegans) |
| chr17_-_39538550 | 0.08 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr5_+_150404904 | 0.08 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr9_-_95298314 | 0.08 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr2_-_46769694 | 0.07 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr10_-_104211294 | 0.07 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr2_-_101925055 | 0.07 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr12_+_57623477 | 0.07 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_+_33865218 | 0.07 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr14_+_102276192 | 0.07 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr5_-_111091948 | 0.07 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr1_+_154378049 | 0.07 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr3_-_196910721 | 0.07 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_+_141103634 | 0.07 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr17_-_685493 | 0.07 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr7_-_130080977 | 0.07 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr8_+_24151553 | 0.06 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr17_-_41623009 | 0.06 |
ENST00000393664.2
|
ETV4
|
ets variant 4 |
| chr13_-_28545276 | 0.06 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr21_+_40824003 | 0.06 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr2_-_99279928 | 0.06 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr6_+_4706368 | 0.06 |
ENST00000328908.5
|
CDYL
|
chromodomain protein, Y-like |
| chr12_+_57624119 | 0.05 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr3_+_158787041 | 0.05 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_-_207143802 | 0.05 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr17_-_41623691 | 0.05 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr8_-_102217515 | 0.05 |
ENST00000520347.1
ENST00000523922.1 ENST00000520984.1 |
ZNF706
|
zinc finger protein 706 |
| chr10_-_97416400 | 0.05 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr1_-_226496898 | 0.05 |
ENST00000481685.1
|
LIN9
|
lin-9 homolog (C. elegans) |
| chr22_-_30867973 | 0.05 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr4_-_85654615 | 0.05 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr7_+_20687017 | 0.05 |
ENST00000258738.6
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr21_+_19617140 | 0.04 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr2_+_191208196 | 0.04 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr3_+_62304712 | 0.03 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr17_-_685559 | 0.03 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr5_-_54830871 | 0.03 |
ENST00000307259.8
|
PPAP2A
|
phosphatidic acid phosphatase type 2A |
| chr20_+_39657454 | 0.03 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr17_-_41322332 | 0.03 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr3_-_48130314 | 0.03 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr14_-_67826486 | 0.03 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
Network of associatons between targets according to the STRING database.
First level regulatory network of HLF_TEF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.5 | 3.6 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 1.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.3 | 0.8 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 2.0 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 1.0 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.3 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 1.8 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.6 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.4 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.5 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 1.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.9 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0030952 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.6 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.3 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.3 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.3 | 0.8 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 3.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.4 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.3 | 1.8 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 2.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 3.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.5 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0052811 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) |
| 0.0 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 1.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 4.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 3.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |