Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
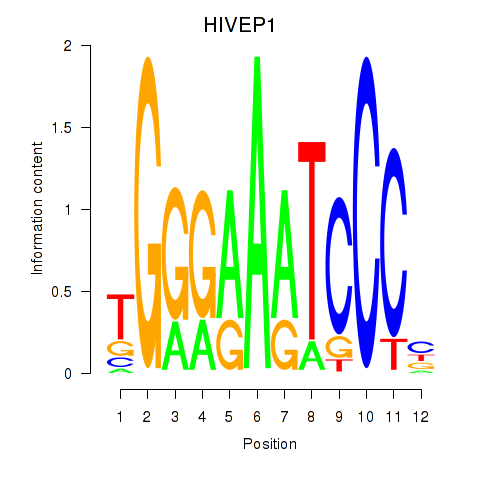
Results for HIVEP1
Z-value: 1.17
Transcription factors associated with HIVEP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIVEP1
|
ENSG00000095951.12 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIVEP1 | hg19_v2_chr6_+_12012170_12012245 | -0.77 | 2.3e-01 | Click! |
Activity profile of HIVEP1 motif
Sorted Z-values of HIVEP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_116595028 | 0.52 |
ENST00000397751.1
|
ST7-OT4
|
ST7 overlapping transcript 4 |
| chr22_+_37257015 | 0.41 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr14_+_52313833 | 0.40 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr19_+_51897742 | 0.36 |
ENST00000600765.1
|
CTD-2616J11.14
|
CTD-2616J11.14 |
| chr3_+_187086120 | 0.33 |
ENST00000259030.2
|
RTP4
|
receptor (chemosensory) transporter protein 4 |
| chr17_-_39093672 | 0.32 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr19_+_52076425 | 0.31 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr19_-_33716750 | 0.28 |
ENST00000253188.4
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chrX_+_48660107 | 0.28 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr6_+_30851205 | 0.28 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_-_99698338 | 0.25 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr10_+_124739911 | 0.25 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr5_+_150591678 | 0.24 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator |
| chr7_+_7606605 | 0.24 |
ENST00000456533.1
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr19_+_35225121 | 0.24 |
ENST00000595708.1
ENST00000593781.1 |
ZNF181
|
zinc finger protein 181 |
| chr22_-_37640456 | 0.24 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr2_-_26251481 | 0.23 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr5_-_150460914 | 0.23 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr17_+_79990058 | 0.23 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr17_+_6918354 | 0.22 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr18_-_2982869 | 0.22 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr19_-_5785630 | 0.22 |
ENST00000586012.1
ENST00000590343.1 |
CTB-54O9.9
DUS3L
|
Uncharacterized protein dihydrouridine synthase 3-like (S. cerevisiae) |
| chr19_-_37178284 | 0.22 |
ENST00000425254.2
ENST00000590952.1 ENST00000433232.1 |
AC074138.3
|
AC074138.3 |
| chr3_-_119396193 | 0.21 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr11_-_119999539 | 0.21 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chr2_-_74781061 | 0.21 |
ENST00000264094.3
ENST00000393937.2 ENST00000409986.1 |
LOXL3
|
lysyl oxidase-like 3 |
| chr22_-_37640277 | 0.21 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr3_-_49893907 | 0.21 |
ENST00000482582.1
|
TRAIP
|
TRAF interacting protein |
| chr11_+_47608198 | 0.21 |
ENST00000356737.2
ENST00000538490.1 |
FAM180B
|
family with sequence similarity 180, member B |
| chr9_-_136344197 | 0.20 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_+_202047843 | 0.20 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr11_-_119999611 | 0.20 |
ENST00000529044.1
|
TRIM29
|
tripartite motif containing 29 |
| chr4_+_86749045 | 0.20 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_+_10365082 | 0.20 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr15_+_75315896 | 0.19 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr19_+_18544045 | 0.19 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr9_-_34381511 | 0.19 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr11_+_45792967 | 0.19 |
ENST00000378779.2
|
CTD-2210P24.4
|
Uncharacterized protein |
| chr12_+_110011571 | 0.19 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr6_-_31508304 | 0.19 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr11_-_17410869 | 0.19 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr19_-_5978090 | 0.18 |
ENST00000592621.1
ENST00000034275.8 ENST00000591092.1 ENST00000591333.1 ENST00000590623.1 ENST00000439268.2 ENST00000587159.1 |
RANBP3
|
RAN binding protein 3 |
| chr17_-_78428487 | 0.18 |
ENST00000562672.2
|
CTD-2526A2.2
|
CTD-2526A2.2 |
| chr5_+_140480083 | 0.18 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr16_+_57392684 | 0.18 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr15_+_74908228 | 0.18 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr16_-_85969774 | 0.18 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr16_+_50730910 | 0.18 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr19_-_46272462 | 0.18 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr6_+_31555045 | 0.18 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr11_+_65292884 | 0.18 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr8_-_65711310 | 0.18 |
ENST00000310193.3
|
CYP7B1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr12_+_64798826 | 0.18 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr11_+_64052294 | 0.17 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_+_156611960 | 0.17 |
ENST00000361588.5
|
BCAN
|
brevican |
| chr10_+_74653330 | 0.17 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr12_+_112451120 | 0.17 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr19_-_17186229 | 0.17 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr19_+_2270283 | 0.17 |
ENST00000588673.2
|
OAZ1
|
ornithine decarboxylase antizyme 1 |
| chr14_+_75988768 | 0.16 |
ENST00000286639.6
|
BATF
|
basic leucine zipper transcription factor, ATF-like |
| chr20_+_1115821 | 0.16 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr2_-_242211359 | 0.16 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr11_+_65292538 | 0.16 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr22_-_37215523 | 0.16 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr20_-_44007014 | 0.16 |
ENST00000372726.3
ENST00000537995.1 |
TP53TG5
|
TP53 target 5 |
| chr1_+_24645915 | 0.16 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_-_39390212 | 0.16 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr1_-_209824643 | 0.16 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr17_-_27044760 | 0.15 |
ENST00000395243.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr5_+_142286887 | 0.15 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr17_+_64961026 | 0.15 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr7_-_100034060 | 0.15 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr15_+_73976715 | 0.15 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr11_-_65314905 | 0.15 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr22_-_29949680 | 0.15 |
ENST00000397873.2
ENST00000490103.1 |
THOC5
|
THO complex 5 |
| chr18_-_72920372 | 0.15 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr1_+_114522049 | 0.15 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr6_+_43484760 | 0.15 |
ENST00000372389.3
ENST00000372344.2 ENST00000304004.3 ENST00000423780.1 |
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa |
| chr9_-_127703333 | 0.14 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr11_+_18287801 | 0.14 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr1_+_46713357 | 0.14 |
ENST00000442598.1
|
RAD54L
|
RAD54-like (S. cerevisiae) |
| chrX_+_47444613 | 0.14 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr12_+_7055631 | 0.14 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_+_86396265 | 0.14 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_+_155051379 | 0.14 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr19_+_45504688 | 0.14 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chrX_-_57147748 | 0.14 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr6_-_33290580 | 0.14 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr22_+_50986462 | 0.14 |
ENST00000395676.2
|
KLHDC7B
|
kelch domain containing 7B |
| chr19_-_10445399 | 0.14 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr7_+_44084262 | 0.14 |
ENST00000456905.1
ENST00000440166.1 ENST00000452943.1 ENST00000468694.1 ENST00000494774.1 ENST00000490734.2 |
DBNL
|
drebrin-like |
| chr11_-_842509 | 0.14 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr19_+_17326141 | 0.14 |
ENST00000445667.2
ENST00000263897.5 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr15_+_74509530 | 0.14 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr1_+_222988464 | 0.14 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr14_+_103243813 | 0.14 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr20_+_2276639 | 0.14 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr19_-_4831701 | 0.14 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chrX_-_48814278 | 0.13 |
ENST00000455452.1
|
OTUD5
|
OTU domain containing 5 |
| chr3_-_124839648 | 0.13 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr19_+_17416609 | 0.13 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr2_-_61108449 | 0.13 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr3_+_57261859 | 0.13 |
ENST00000495803.1
ENST00000444459.1 |
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr19_-_55628927 | 0.13 |
ENST00000263433.3
ENST00000376393.2 |
PPP1R12C
|
protein phosphatase 1, regulatory subunit 12C |
| chr6_-_33256664 | 0.13 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr7_+_150497491 | 0.13 |
ENST00000484928.1
|
TMEM176A
|
transmembrane protein 176A |
| chr4_+_86396321 | 0.13 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr19_-_41220540 | 0.13 |
ENST00000594490.1
|
ADCK4
|
aarF domain containing kinase 4 |
| chr1_+_24645865 | 0.13 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_+_6918093 | 0.13 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr17_-_18908040 | 0.13 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr19_+_41869894 | 0.13 |
ENST00000413014.2
|
TMEM91
|
transmembrane protein 91 |
| chr15_+_74908147 | 0.13 |
ENST00000568139.1
ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3
|
CDC-like kinase 3 |
| chr19_+_17326191 | 0.13 |
ENST00000595101.1
ENST00000596136.1 ENST00000379776.4 |
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr17_-_79917645 | 0.13 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr6_+_138188351 | 0.12 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_-_64570706 | 0.12 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr1_+_230193521 | 0.12 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr1_+_24645807 | 0.12 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr6_+_29691198 | 0.12 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr10_-_118031778 | 0.12 |
ENST00000369236.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chrX_-_73072534 | 0.12 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr17_+_7461781 | 0.12 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_1846519 | 0.12 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr7_+_100271446 | 0.12 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr16_-_12070468 | 0.12 |
ENST00000538896.1
|
RP11-166B2.1
|
Putative NPIP-like protein LOC729978 |
| chr14_-_23292596 | 0.12 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_-_9291063 | 0.12 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr1_+_156611900 | 0.12 |
ENST00000457777.2
ENST00000424639.1 |
BCAN
|
brevican |
| chr22_-_50708781 | 0.12 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr20_+_327668 | 0.12 |
ENST00000382291.3
ENST00000609504.1 ENST00000382285.2 |
NRSN2
|
neurensin 2 |
| chr7_+_69064300 | 0.12 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr7_-_149158187 | 0.12 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr17_+_35306175 | 0.11 |
ENST00000225402.5
|
AATF
|
apoptosis antagonizing transcription factor |
| chr22_-_29949634 | 0.11 |
ENST00000397872.1
ENST00000397871.1 ENST00000440771.1 |
THOC5
|
THO complex 5 |
| chr1_+_111770278 | 0.11 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr1_-_2706236 | 0.11 |
ENST00000401095.3
ENST00000574621.2 |
TTC34
|
tetratricopeptide repeat domain 34 |
| chr19_-_40336969 | 0.11 |
ENST00000599134.1
ENST00000597634.1 ENST00000598417.1 ENST00000601274.1 ENST00000594309.1 ENST00000221801.3 |
FBL
|
fibrillarin |
| chr5_-_150460539 | 0.11 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr16_+_88772866 | 0.11 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr17_+_38024417 | 0.11 |
ENST00000348931.4
ENST00000583811.1 ENST00000584588.1 ENST00000377940.3 |
ZPBP2
|
zona pellucida binding protein 2 |
| chr17_+_40440481 | 0.11 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr12_+_53342625 | 0.11 |
ENST00000388837.2
ENST00000550600.1 ENST00000388835.3 |
KRT18
|
keratin 18 |
| chr2_+_3622932 | 0.11 |
ENST00000406376.1
|
RPS7
|
ribosomal protein S7 |
| chr17_-_34207295 | 0.11 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr19_+_54694119 | 0.11 |
ENST00000456872.1
ENST00000302937.4 ENST00000429671.2 |
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr9_-_34381536 | 0.11 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr19_-_4182530 | 0.11 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr17_+_42977122 | 0.11 |
ENST00000412523.2
ENST00000331733.4 ENST00000417826.2 |
FAM187A
CCDC103
|
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr17_+_43209967 | 0.11 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr17_+_6918064 | 0.11 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr1_+_46713404 | 0.11 |
ENST00000371975.4
ENST00000469835.1 |
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr7_-_99097863 | 0.11 |
ENST00000426306.2
ENST00000337673.6 |
ZNF394
|
zinc finger protein 394 |
| chr1_+_222988406 | 0.11 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr17_+_37793318 | 0.10 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr14_-_69446034 | 0.10 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr19_+_14672755 | 0.10 |
ENST00000594545.1
|
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr19_+_39390587 | 0.10 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr22_-_19512893 | 0.10 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr1_-_206671061 | 0.10 |
ENST00000367119.1
|
C1orf147
|
chromosome 1 open reading frame 147 |
| chr17_-_74236382 | 0.10 |
ENST00000592271.1
ENST00000319945.6 ENST00000269391.6 |
RNF157
|
ring finger protein 157 |
| chr11_+_64052692 | 0.10 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr16_+_75032901 | 0.10 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr11_+_66247880 | 0.10 |
ENST00000360510.2
ENST00000453114.1 ENST00000541961.1 ENST00000532019.1 ENST00000526515.1 ENST00000530165.1 ENST00000533725.1 |
DPP3
|
dipeptidyl-peptidase 3 |
| chr15_+_91411810 | 0.10 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr3_+_52280173 | 0.10 |
ENST00000296487.4
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr4_-_80994210 | 0.10 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr5_-_138862326 | 0.10 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr19_-_46366392 | 0.10 |
ENST00000598059.1
ENST00000594293.1 ENST00000245934.7 |
SYMPK
|
symplekin |
| chr1_+_161195781 | 0.10 |
ENST00000367988.3
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr3_+_57261743 | 0.10 |
ENST00000288266.3
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr17_-_27045405 | 0.10 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr12_-_58131931 | 0.10 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_+_29910301 | 0.10 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr16_+_28722809 | 0.10 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr17_-_9479128 | 0.10 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr3_+_186353756 | 0.10 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr1_+_45477819 | 0.10 |
ENST00000246337.4
|
UROD
|
uroporphyrinogen decarboxylase |
| chr5_+_140552218 | 0.10 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr6_-_31239846 | 0.09 |
ENST00000415537.1
ENST00000376228.5 ENST00000383329.3 |
HLA-C
|
major histocompatibility complex, class I, C |
| chr2_+_234686976 | 0.09 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr11_+_18287721 | 0.09 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr12_+_53662073 | 0.09 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr20_+_44746939 | 0.09 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr11_-_67276100 | 0.09 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr19_-_2042065 | 0.09 |
ENST00000591588.1
ENST00000591142.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr7_-_132261253 | 0.09 |
ENST00000321063.4
|
PLXNA4
|
plexin A4 |
| chr17_+_37793378 | 0.09 |
ENST00000544210.2
ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr1_+_37940153 | 0.09 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr10_-_93669233 | 0.09 |
ENST00000311575.5
|
FGFBP3
|
fibroblast growth factor binding protein 3 |
| chr19_+_41256764 | 0.09 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr12_+_7053172 | 0.09 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr17_-_56605341 | 0.09 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr6_-_10694766 | 0.09 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr22_-_50964849 | 0.09 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr12_+_53662110 | 0.09 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr6_+_33378517 | 0.09 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr2_+_74154032 | 0.09 |
ENST00000356837.6
|
DGUOK
|
deoxyguanosine kinase |
| chr19_+_18496957 | 0.09 |
ENST00000252809.3
|
GDF15
|
growth differentiation factor 15 |
| chr19_+_35739897 | 0.09 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIVEP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.3 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.3 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.5 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.1 | GO:2000625 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:1902462 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) activation of protein kinase C activity(GO:1990051) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0048298 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |