Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
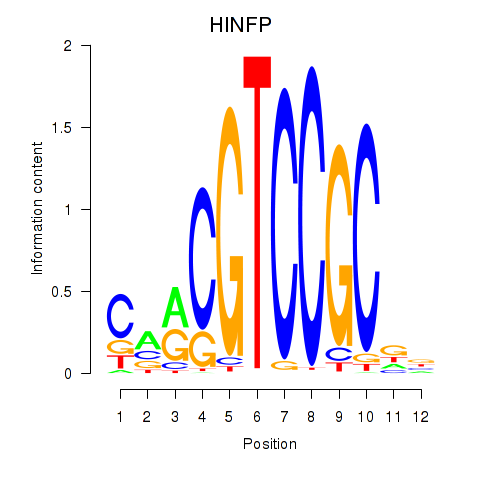
Results for HINFP
Z-value: 0.56
Transcription factors associated with HINFP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HINFP
|
ENSG00000172273.8 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HINFP | hg19_v2_chr11_+_118992269_118992334 | 0.96 | 4.3e-02 | Click! |
Activity profile of HINFP motif
Sorted Z-values of HINFP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54962625 | 0.38 |
ENST00000559432.1
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr3_-_57583185 | 0.33 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr16_-_54962415 | 0.33 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr2_+_64068116 | 0.31 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr1_-_51796226 | 0.24 |
ENST00000451380.1
ENST00000371747.3 ENST00000439482.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr14_+_75894391 | 0.24 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr2_+_28974489 | 0.23 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr13_-_48877795 | 0.22 |
ENST00000436963.1
ENST00000433480.2 |
LINC00441
|
long intergenic non-protein coding RNA 441 |
| chr12_+_107349606 | 0.21 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr22_+_29168652 | 0.21 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr11_+_3877412 | 0.19 |
ENST00000527651.1
|
STIM1
|
stromal interaction molecule 1 |
| chr17_+_30593195 | 0.18 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr3_+_110790590 | 0.18 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr13_-_99738867 | 0.18 |
ENST00000427887.1
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr5_-_141257954 | 0.17 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr17_-_42276574 | 0.17 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr16_-_54962704 | 0.17 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr12_-_117175819 | 0.16 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr12_+_56522001 | 0.16 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr17_-_28619059 | 0.16 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr17_+_74380683 | 0.15 |
ENST00000592299.1
ENST00000590959.1 ENST00000323374.4 |
SPHK1
|
sphingosine kinase 1 |
| chr3_-_49449350 | 0.15 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr12_+_53773944 | 0.15 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr17_-_1419914 | 0.14 |
ENST00000397335.3
ENST00000574561.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr12_+_49760639 | 0.14 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chrX_-_153285395 | 0.14 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr12_-_90103077 | 0.13 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr11_-_66336060 | 0.13 |
ENST00000310325.5
|
CTSF
|
cathepsin F |
| chr5_+_61601965 | 0.13 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chrX_-_153285251 | 0.13 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr3_+_183853052 | 0.13 |
ENST00000273783.3
ENST00000432569.1 ENST00000444495.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa |
| chr17_+_46125685 | 0.12 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr3_-_113415441 | 0.12 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr17_-_46690839 | 0.12 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr5_+_61602055 | 0.12 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr14_+_75894714 | 0.12 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr17_+_43971643 | 0.11 |
ENST00000344290.5
ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT
|
microtubule-associated protein tau |
| chr13_+_111805980 | 0.11 |
ENST00000491775.1
ENST00000466143.1 ENST00000544132.1 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr14_-_101034811 | 0.11 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr6_-_33267101 | 0.11 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr8_-_101733794 | 0.11 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_+_62104897 | 0.11 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr2_+_204193129 | 0.11 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr1_-_36915880 | 0.11 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr7_-_29234802 | 0.11 |
ENST00000449801.1
ENST00000409850.1 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr7_-_134143841 | 0.11 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr2_+_64068074 | 0.10 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr12_+_69633407 | 0.10 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chrX_-_153602991 | 0.10 |
ENST00000369850.3
ENST00000422373.1 |
FLNA
|
filamin A, alpha |
| chr12_+_56137064 | 0.10 |
ENST00000257868.5
ENST00000546799.1 |
GDF11
|
growth differentiation factor 11 |
| chr16_+_84002234 | 0.10 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr3_+_196295482 | 0.10 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr1_+_28995231 | 0.10 |
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr17_+_74381343 | 0.10 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr9_-_94186131 | 0.09 |
ENST00000297689.3
|
NFIL3
|
nuclear factor, interleukin 3 regulated |
| chr10_+_94608245 | 0.09 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr6_+_116692102 | 0.09 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase |
| chr17_-_48450265 | 0.09 |
ENST00000507088.1
|
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr4_-_156298028 | 0.09 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr19_+_36545781 | 0.09 |
ENST00000388999.3
|
WDR62
|
WD repeat domain 62 |
| chr2_-_33824336 | 0.09 |
ENST00000431950.1
ENST00000403368.1 ENST00000441530.2 |
FAM98A
|
family with sequence similarity 98, member A |
| chr17_-_1419878 | 0.09 |
ENST00000449479.1
ENST00000477910.1 ENST00000542125.1 ENST00000575172.1 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr2_+_204193149 | 0.09 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr6_+_56819895 | 0.08 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr8_+_6565854 | 0.08 |
ENST00000285518.6
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr6_+_35310391 | 0.08 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr6_-_33266687 | 0.08 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr7_+_128095900 | 0.08 |
ENST00000435296.2
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr1_+_109102652 | 0.08 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr4_+_148402069 | 0.08 |
ENST00000358556.4
ENST00000339690.5 ENST00000511804.1 ENST00000324300.5 |
EDNRA
|
endothelin receptor type A |
| chr1_-_36916066 | 0.08 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr7_+_65540853 | 0.08 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr3_-_48936272 | 0.08 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr19_+_48112371 | 0.08 |
ENST00000594866.1
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1 |
| chr14_+_52118694 | 0.08 |
ENST00000554778.1
|
FRMD6
|
FERM domain containing 6 |
| chr3_+_50654821 | 0.08 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr7_+_128095945 | 0.07 |
ENST00000257696.4
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr17_-_1420006 | 0.07 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr7_-_117513540 | 0.07 |
ENST00000160373.3
|
CTTNBP2
|
cortactin binding protein 2 |
| chr14_-_23504087 | 0.07 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr4_+_166128836 | 0.07 |
ENST00000511305.1
|
KLHL2
|
kelch-like family member 2 |
| chr9_-_74383302 | 0.07 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr12_-_90102594 | 0.07 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr3_-_13009168 | 0.07 |
ENST00000273221.4
|
IQSEC1
|
IQ motif and Sec7 domain 1 |
| chr13_-_111214015 | 0.07 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chrX_+_71996972 | 0.07 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr1_-_36916011 | 0.07 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr12_+_49761147 | 0.07 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr14_+_24769043 | 0.07 |
ENST00000267425.3
ENST00000396802.3 |
NOP9
|
NOP9 nucleolar protein |
| chr11_-_73309112 | 0.07 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr7_+_156931606 | 0.07 |
ENST00000348165.5
|
UBE3C
|
ubiquitin protein ligase E3C |
| chr12_+_56521840 | 0.07 |
ENST00000394048.5
|
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr17_-_7232585 | 0.07 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr16_-_2162831 | 0.07 |
ENST00000483024.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr22_-_43010859 | 0.07 |
ENST00000339677.6
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr3_-_52719810 | 0.07 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr8_+_110656344 | 0.07 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr20_+_42295745 | 0.06 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr11_+_66045634 | 0.06 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr7_+_100271446 | 0.06 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr15_+_101459420 | 0.06 |
ENST00000388948.3
ENST00000284395.5 ENST00000534045.1 ENST00000532029.2 |
LRRK1
|
leucine-rich repeat kinase 1 |
| chr1_+_39957292 | 0.06 |
ENST00000331593.5
|
BMP8A
|
bone morphogenetic protein 8a |
| chr4_-_77997126 | 0.06 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr4_-_2758015 | 0.06 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr22_+_32871224 | 0.06 |
ENST00000452138.1
ENST00000382058.3 ENST00000397426.1 |
FBXO7
|
F-box protein 7 |
| chr6_-_24721054 | 0.06 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr11_-_73309228 | 0.06 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr9_-_131085021 | 0.06 |
ENST00000372890.4
|
TRUB2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr16_+_2510081 | 0.06 |
ENST00000361837.4
ENST00000569496.1 ENST00000567489.1 ENST00000563531.1 ENST00000483320.1 |
C16orf59
|
chromosome 16 open reading frame 59 |
| chr4_-_183838747 | 0.06 |
ENST00000438320.2
|
DCTD
|
dCMP deaminase |
| chr7_+_98972327 | 0.06 |
ENST00000455009.1
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr9_+_96214166 | 0.06 |
ENST00000375389.3
ENST00000333936.5 ENST00000340893.4 |
FAM120A
|
family with sequence similarity 120A |
| chr3_-_38691119 | 0.06 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr3_+_49209023 | 0.06 |
ENST00000332780.2
|
KLHDC8B
|
kelch domain containing 8B |
| chr5_-_158526756 | 0.06 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr13_+_21714913 | 0.06 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr19_+_34972543 | 0.06 |
ENST00000590071.2
|
WTIP
|
Wilms tumor 1 interacting protein |
| chr14_-_24768913 | 0.06 |
ENST00000288111.7
|
DHRS1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr9_+_139921916 | 0.06 |
ENST00000314330.2
|
C9orf139
|
chromosome 9 open reading frame 139 |
| chr6_-_139695757 | 0.06 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr7_-_76256557 | 0.06 |
ENST00000275569.4
ENST00000310842.4 |
POMZP3
|
POM121 and ZP3 fusion |
| chr5_-_158526693 | 0.06 |
ENST00000380654.4
|
EBF1
|
early B-cell factor 1 |
| chr17_-_7518145 | 0.06 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr7_+_100271355 | 0.05 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr2_+_121010370 | 0.05 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr6_+_26104104 | 0.05 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr9_+_100174232 | 0.05 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr10_-_135187193 | 0.05 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr17_+_46125707 | 0.05 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr10_+_75532028 | 0.05 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr2_+_30670209 | 0.05 |
ENST00000497423.1
ENST00000476535.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr17_+_27895045 | 0.05 |
ENST00000580183.2
ENST00000578749.1 ENST00000582829.2 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr17_-_48450534 | 0.05 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr8_-_102218292 | 0.05 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr9_+_84304628 | 0.05 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr11_+_125462690 | 0.05 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr9_+_100174344 | 0.05 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr12_-_99288536 | 0.05 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_+_268148 | 0.05 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr7_-_107643567 | 0.05 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr8_+_32405728 | 0.05 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr17_-_1420182 | 0.05 |
ENST00000421807.2
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr5_-_132166303 | 0.05 |
ENST00000440118.1
|
SHROOM1
|
shroom family member 1 |
| chr22_+_32870661 | 0.05 |
ENST00000266087.7
|
FBXO7
|
F-box protein 7 |
| chr2_+_120301997 | 0.05 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr22_-_28197486 | 0.05 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr20_+_30458431 | 0.05 |
ENST00000375938.4
ENST00000535842.1 ENST00000310998.4 ENST00000375921.2 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr5_+_142149932 | 0.05 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_+_171785824 | 0.05 |
ENST00000452526.2
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa |
| chr20_-_30458354 | 0.05 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr1_-_22109484 | 0.05 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr20_+_3451732 | 0.05 |
ENST00000446916.2
|
ATRN
|
attractin |
| chr3_-_49761337 | 0.04 |
ENST00000535833.1
ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3
GMPPB
|
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr6_+_35310312 | 0.04 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr10_-_128994422 | 0.04 |
ENST00000522781.1
|
FAM196A
|
family with sequence similarity 196, member A |
| chr22_-_43010928 | 0.04 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr17_+_46126135 | 0.04 |
ENST00000361665.3
ENST00000585062.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr13_-_48612067 | 0.04 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr8_-_100905850 | 0.04 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr14_-_99947121 | 0.04 |
ENST00000329331.3
ENST00000436070.2 |
SETD3
|
SET domain containing 3 |
| chr7_+_65540780 | 0.04 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chr15_-_35280426 | 0.04 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr1_+_212208919 | 0.04 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr8_-_142318398 | 0.04 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr9_+_37120536 | 0.04 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr2_+_120770581 | 0.04 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr11_-_75062730 | 0.04 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr5_-_136834982 | 0.04 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr17_+_17206635 | 0.04 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr10_+_99079008 | 0.04 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr17_+_48450575 | 0.04 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr4_-_183838482 | 0.04 |
ENST00000357067.3
ENST00000510370.1 |
DCTD
|
dCMP deaminase |
| chr17_+_12693001 | 0.04 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr17_-_27621125 | 0.04 |
ENST00000579665.1
ENST00000225388.4 |
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr8_-_27462822 | 0.04 |
ENST00000522098.1
|
CLU
|
clusterin |
| chrX_+_154299690 | 0.04 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr8_+_32405785 | 0.03 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr5_-_141030943 | 0.03 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr19_-_39881669 | 0.03 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr11_+_64073699 | 0.03 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr8_+_26240414 | 0.03 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr20_+_3451650 | 0.03 |
ENST00000262919.5
|
ATRN
|
attractin |
| chr12_-_1703331 | 0.03 |
ENST00000339235.3
|
FBXL14
|
F-box and leucine-rich repeat protein 14 |
| chr1_+_22351977 | 0.03 |
ENST00000420503.1
ENST00000416769.1 ENST00000404210.2 |
LINC00339
|
long intergenic non-protein coding RNA 339 |
| chr11_+_65819802 | 0.03 |
ENST00000528302.1
ENST00000322535.6 ENST00000524627.1 ENST00000533595.1 ENST00000530322.1 |
SF3B2
|
splicing factor 3b, subunit 2, 145kDa |
| chr3_-_57583130 | 0.03 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr7_+_101459263 | 0.03 |
ENST00000292538.4
ENST00000393824.3 ENST00000547394.2 ENST00000360264.3 ENST00000425244.2 |
CUX1
|
cut-like homeobox 1 |
| chr4_-_183838372 | 0.03 |
ENST00000503820.1
ENST00000503988.1 |
DCTD
|
dCMP deaminase |
| chr8_-_100905363 | 0.03 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr1_+_204797749 | 0.03 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chr6_+_42847348 | 0.03 |
ENST00000493763.1
ENST00000304734.5 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr6_+_159291090 | 0.03 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr5_-_159546396 | 0.03 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr7_+_97910981 | 0.03 |
ENST00000297290.3
|
BRI3
|
brain protein I3 |
| chr17_-_42402138 | 0.03 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr1_-_2706236 | 0.03 |
ENST00000401095.3
ENST00000574621.2 |
TTC34
|
tetratricopeptide repeat domain 34 |
| chr5_+_14143728 | 0.03 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr15_+_91643442 | 0.03 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr15_-_75932528 | 0.03 |
ENST00000403490.1
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr1_-_197115818 | 0.03 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
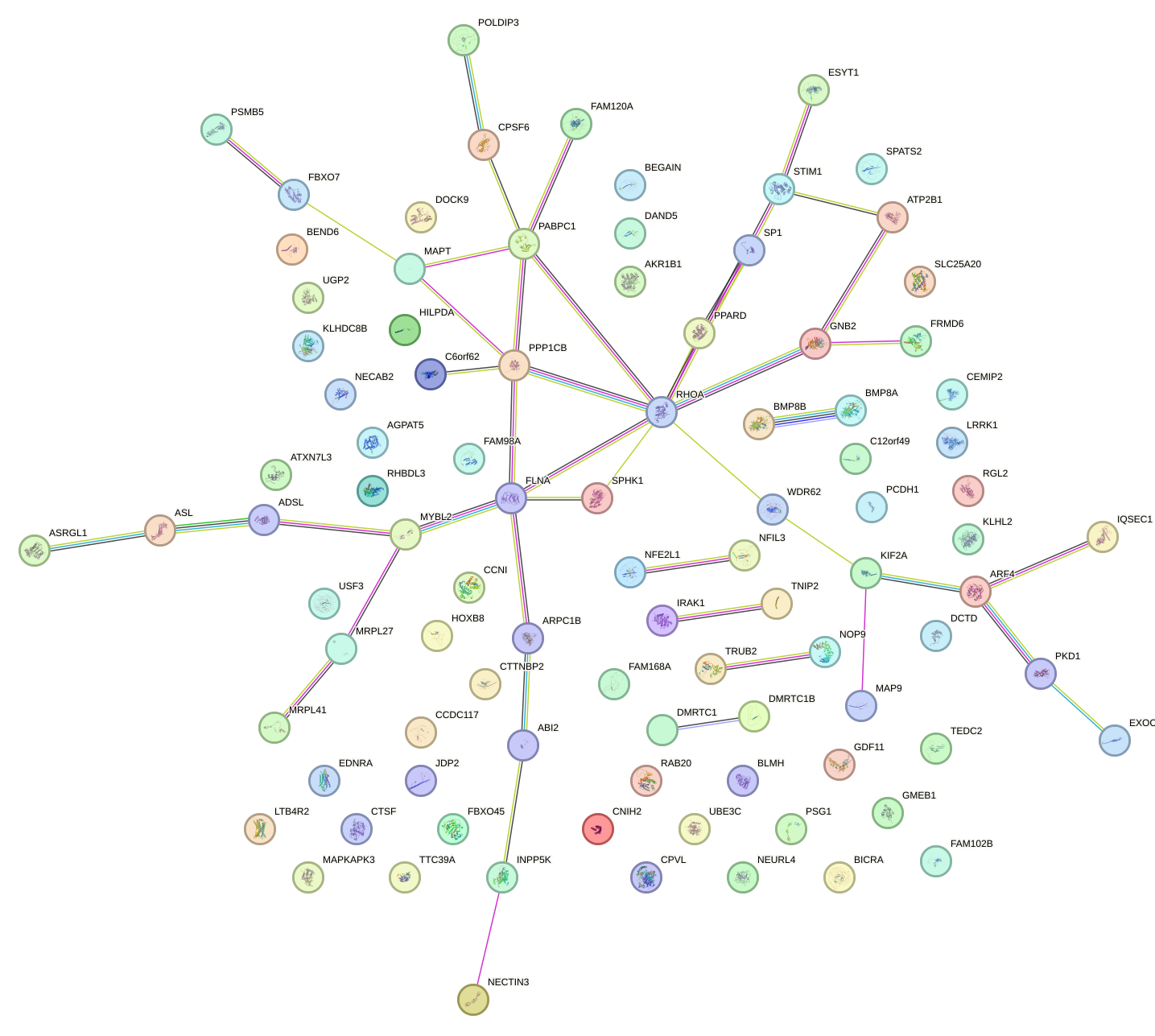
Network of associatons between targets according to the STRING database.
First level regulatory network of HINFP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.3 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.1 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.0 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.0 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0070814 | phenotypic switching(GO:0036166) hydrogen sulfide biosynthetic process(GO:0070814) regulation of phenotypic switching(GO:1900239) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |