Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
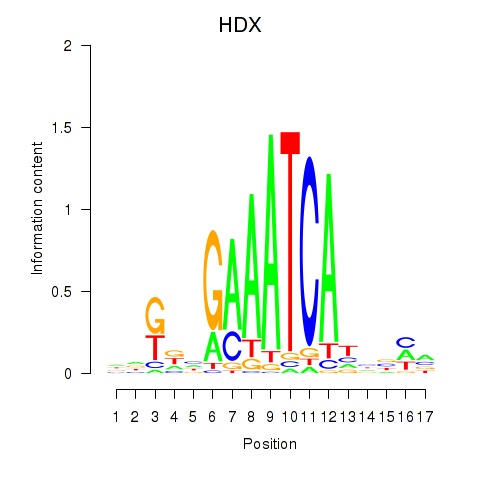
Results for HDX
Z-value: 0.34
Transcription factors associated with HDX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HDX
|
ENSG00000165259.9 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HDX | hg19_v2_chrX_-_83757399_83757487 | 0.61 | 3.9e-01 | Click! |
Activity profile of HDX motif
Sorted Z-values of HDX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_139756791 | 0.60 |
ENST00000489809.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr6_+_144471643 | 0.30 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chrX_+_100224676 | 0.29 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr12_+_10460417 | 0.19 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr2_+_169926047 | 0.15 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr2_+_149447783 | 0.11 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr2_+_87754887 | 0.11 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr8_+_145065705 | 0.10 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr5_-_35230771 | 0.09 |
ENST00000342362.5
|
PRLR
|
prolactin receptor |
| chr10_-_21186144 | 0.09 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr14_+_79746249 | 0.07 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr14_+_71788096 | 0.07 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr5_-_135290651 | 0.06 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr14_+_79745746 | 0.06 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr10_-_14372870 | 0.06 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr5_-_135290705 | 0.06 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_35230649 | 0.06 |
ENST00000382002.5
|
PRLR
|
prolactin receptor |
| chr7_-_107883678 | 0.06 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr6_+_42584847 | 0.05 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr19_-_58485895 | 0.05 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr3_+_54157480 | 0.04 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr19_+_36602104 | 0.04 |
ENST00000585332.1
ENST00000262637.4 |
OVOL3
|
ovo-like zinc finger 3 |
| chr9_+_6215799 | 0.04 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr9_+_35538616 | 0.04 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr1_-_159684371 | 0.04 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr1_-_11024258 | 0.04 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr2_+_102758210 | 0.03 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr2_-_40657397 | 0.03 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr3_-_100565249 | 0.03 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_-_48866517 | 0.02 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr10_+_45406627 | 0.02 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr14_+_79745682 | 0.02 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr1_+_75600567 | 0.02 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr12_-_31743901 | 0.02 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr5_-_41213607 | 0.01 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr1_-_169680745 | 0.01 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr1_+_47489240 | 0.01 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr1_+_84630352 | 0.01 |
ENST00000450730.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chrX_+_27608490 | 0.01 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr7_-_135412925 | 0.01 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr8_+_24151620 | 0.01 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr7_+_144015218 | 0.01 |
ENST00000408951.1
|
OR2A1
|
olfactory receptor, family 2, subfamily A, member 1 |
| chr2_+_128403720 | 0.01 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr13_+_24144796 | 0.00 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr18_+_29027696 | 0.00 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HDX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |