Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
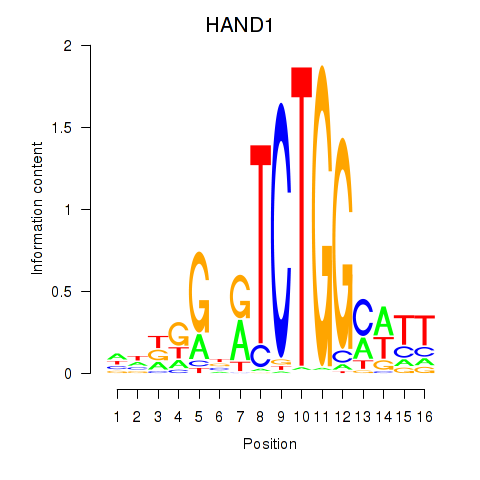
Results for HAND1
Z-value: 0.86
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.2 | heart and neural crest derivatives expressed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HAND1 | hg19_v2_chr5_-_153857819_153857824 | -0.25 | 7.5e-01 | Click! |
Activity profile of HAND1 motif
Sorted Z-values of HAND1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_64216748 | 1.19 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr19_-_36304201 | 0.76 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr2_+_13677795 | 0.59 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr7_+_76139833 | 0.58 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr3_-_114035026 | 0.58 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr3_-_20227619 | 0.57 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr1_+_28261492 | 0.55 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_-_28621353 | 0.53 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr14_+_93673574 | 0.51 |
ENST00000554232.1
ENST00000556871.1 ENST00000555113.1 |
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr6_+_80341000 | 0.50 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr1_+_28261621 | 0.50 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_-_10652993 | 0.50 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr10_+_99344071 | 0.50 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr12_+_52450298 | 0.48 |
ENST00000550582.2
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr11_+_101785727 | 0.45 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr17_+_6347761 | 0.45 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr14_-_65409502 | 0.41 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr14_-_65409438 | 0.39 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr10_-_118032697 | 0.38 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr1_+_28261533 | 0.38 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr3_-_15382875 | 0.36 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr6_-_133119668 | 0.35 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr19_+_47421933 | 0.34 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr12_+_49717019 | 0.33 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr11_+_124933191 | 0.32 |
ENST00000532000.1
ENST00000308074.4 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr5_-_41870621 | 0.31 |
ENST00000196371.5
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr16_-_4303767 | 0.30 |
ENST00000573268.1
ENST00000573042.1 |
RP11-95P2.1
|
RP11-95P2.1 |
| chr20_+_3713314 | 0.30 |
ENST00000254963.2
ENST00000542646.1 ENST00000399701.1 |
HSPA12B
|
heat shock 70kD protein 12B |
| chr1_+_155911480 | 0.30 |
ENST00000368318.3
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4 |
| chr11_+_124932986 | 0.30 |
ENST00000407458.1
ENST00000298280.5 |
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr3_-_20227720 | 0.30 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr6_+_44191507 | 0.29 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr15_-_33447055 | 0.27 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr17_-_43025005 | 0.27 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr1_-_205601064 | 0.27 |
ENST00000357992.4
ENST00000289703.4 |
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1) |
| chr12_+_53848505 | 0.25 |
ENST00000552819.1
ENST00000455667.3 |
PCBP2
|
poly(rC) binding protein 2 |
| chr1_+_27719148 | 0.25 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr3_+_148415571 | 0.24 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr4_-_25161996 | 0.23 |
ENST00000513285.1
ENST00000382103.2 |
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr17_+_6347729 | 0.23 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr9_-_115095229 | 0.22 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr1_-_155904161 | 0.21 |
ENST00000368319.3
|
KIAA0907
|
KIAA0907 |
| chr11_+_124932955 | 0.21 |
ENST00000403796.2
|
SLC37A2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr19_-_55866061 | 0.20 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr19_+_39647271 | 0.20 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr2_-_228582709 | 0.20 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr2_-_197791441 | 0.20 |
ENST00000409475.1
ENST00000354764.4 ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins 1 |
| chr1_-_155904187 | 0.20 |
ENST00000368321.3
ENST00000368320.3 |
KIAA0907
|
KIAA0907 |
| chr12_-_7261772 | 0.19 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr22_+_31518938 | 0.19 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr12_+_53848549 | 0.19 |
ENST00000439930.3
ENST00000548933.1 ENST00000562264.1 |
PCBP2
|
poly(rC) binding protein 2 |
| chr17_+_46133307 | 0.19 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr18_+_55712915 | 0.18 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr16_-_28621298 | 0.18 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr10_-_118032979 | 0.17 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr1_+_215747118 | 0.17 |
ENST00000448333.1
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr17_+_45908974 | 0.17 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr18_+_12407895 | 0.17 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr4_-_25162188 | 0.16 |
ENST00000302922.3
|
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr1_+_164528866 | 0.16 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chrX_-_34675391 | 0.16 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr20_+_816695 | 0.16 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr15_-_76304731 | 0.14 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr20_+_34020827 | 0.14 |
ENST00000374375.1
|
GDF5OS
|
growth differentiation factor 5 opposite strand |
| chr2_-_42588338 | 0.13 |
ENST00000234301.2
|
COX7A2L
|
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr6_-_117923610 | 0.13 |
ENST00000535237.1
ENST00000052569.6 |
GOPC
|
golgi-associated PDZ and coiled-coil motif containing |
| chr1_-_223988426 | 0.13 |
ENST00000391879.2
|
TP53BP2
|
tumor protein p53 binding protein, 2 |
| chr9_-_115095123 | 0.13 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr16_-_28621312 | 0.13 |
ENST00000314752.7
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_40631859 | 0.12 |
ENST00000295971.7
ENST00000319592.4 |
RBM47
|
RNA binding motif protein 47 |
| chr6_-_39282221 | 0.12 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr3_-_9811595 | 0.12 |
ENST00000256460.3
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr16_-_28608364 | 0.12 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr12_+_98909260 | 0.11 |
ENST00000556029.1
|
TMPO
|
thymopoietin |
| chr16_+_56969284 | 0.11 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr9_-_35828576 | 0.11 |
ENST00000377984.2
ENST00000423537.2 ENST00000472182.1 |
FAM221B
|
family with sequence similarity 221, member B |
| chr22_-_24093267 | 0.11 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70 |
| chr17_+_53342311 | 0.11 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr12_+_68042495 | 0.10 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr11_+_67171358 | 0.10 |
ENST00000526387.1
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr11_-_72432950 | 0.10 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_-_52344416 | 0.10 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr6_-_53013620 | 0.10 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr5_+_167913450 | 0.10 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr1_+_110036699 | 0.10 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr11_-_62476694 | 0.10 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr2_+_3642545 | 0.10 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chr16_-_28608424 | 0.10 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr7_-_140714430 | 0.10 |
ENST00000393008.3
|
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr17_+_75452447 | 0.09 |
ENST00000591472.2
|
SEPT9
|
septin 9 |
| chr21_+_34602377 | 0.09 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr4_-_40632605 | 0.09 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr7_-_142583506 | 0.08 |
ENST00000359396.3
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr4_+_76481258 | 0.08 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chrX_-_133930285 | 0.08 |
ENST00000486347.1
ENST00000343004.5 |
FAM122B
|
family with sequence similarity 122B |
| chr19_+_45165545 | 0.08 |
ENST00000592472.1
ENST00000587729.1 ENST00000585657.1 ENST00000592789.1 ENST00000591979.1 |
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr12_+_68042517 | 0.07 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr2_-_159237472 | 0.07 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr7_+_141490017 | 0.07 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr4_+_22999152 | 0.07 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr10_+_99344104 | 0.07 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr11_+_67171548 | 0.07 |
ENST00000542590.1
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr17_-_45908875 | 0.06 |
ENST00000351111.2
ENST00000414011.1 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr7_-_140714739 | 0.06 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr1_+_174846570 | 0.06 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_+_110036728 | 0.05 |
ENST00000369868.3
ENST00000430195.2 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr4_+_41614909 | 0.05 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_+_67663056 | 0.05 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr12_+_54379569 | 0.05 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr19_-_37701386 | 0.05 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chr4_-_40632757 | 0.04 |
ENST00000511902.1
ENST00000505220.1 |
RBM47
|
RNA binding motif protein 47 |
| chr17_+_66624280 | 0.04 |
ENST00000585484.1
|
RP11-118B18.1
|
RP11-118B18.1 |
| chr3_-_43147431 | 0.04 |
ENST00000441964.1
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr14_+_24422795 | 0.04 |
ENST00000313250.5
ENST00000558263.1 ENST00000543741.2 ENST00000421831.1 ENST00000397073.2 ENST00000308178.8 ENST00000382761.3 ENST00000397075.3 ENST00000397074.3 ENST00000559632.1 ENST00000558581.1 |
DHRS4
|
dehydrogenase/reductase (SDR family) member 4 |
| chrX_+_149531606 | 0.04 |
ENST00000432680.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chr7_+_140378955 | 0.04 |
ENST00000473512.1
|
ADCK2
|
aarF domain containing kinase 2 |
| chr19_+_12862604 | 0.03 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr12_-_110271178 | 0.03 |
ENST00000261740.2
ENST00000392719.2 ENST00000346520.2 |
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr12_+_26111823 | 0.03 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr11_+_67171391 | 0.03 |
ENST00000312390.5
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr17_-_40337470 | 0.03 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr3_+_46919235 | 0.03 |
ENST00000449590.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr17_-_34890037 | 0.03 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr3_-_43147549 | 0.02 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr3_+_141103634 | 0.02 |
ENST00000507722.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_-_81399355 | 0.02 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399411 | 0.02 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399329 | 0.01 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_+_202091980 | 0.01 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HAND1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.5 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 1.0 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.6 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 1.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.8 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |