Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GUGCAAA
Z-value: 0.82
miRNA associated with seed GUGCAAA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-19a-3p
|
MIMAT0000073 |
|
hsa-miR-19b-3p
|
MIMAT0000074 |
Activity profile of GUGCAAA motif
Sorted Z-values of GUGCAAA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_9136758 | 0.30 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr12_+_79258547 | 0.30 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr15_+_98503922 | 0.29 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr1_-_244615425 | 0.28 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chrX_-_135056216 | 0.28 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chrX_+_117480036 | 0.28 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr15_+_52311398 | 0.27 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr8_-_103876965 | 0.27 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chr1_-_51984908 | 0.26 |
ENST00000371730.2
|
EPS15
|
epidermal growth factor receptor pathway substrate 15 |
| chr15_+_78730531 | 0.24 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr1_-_235491462 | 0.24 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr1_-_185286461 | 0.23 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr4_+_57774042 | 0.22 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr20_-_52210368 | 0.22 |
ENST00000371471.2
|
ZNF217
|
zinc finger protein 217 |
| chr8_+_59323823 | 0.22 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr8_+_104311059 | 0.21 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chr1_+_26438289 | 0.20 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr1_-_93426998 | 0.19 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr3_+_30648066 | 0.19 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr8_+_6565854 | 0.19 |
ENST00000285518.6
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr20_-_23402028 | 0.19 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr5_+_78908233 | 0.18 |
ENST00000453514.1
ENST00000423041.2 ENST00000504233.1 ENST00000428308.2 |
PAPD4
|
PAP associated domain containing 4 |
| chr5_-_73937244 | 0.18 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr8_+_87354945 | 0.18 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr4_-_77819002 | 0.18 |
ENST00000334306.2
|
SOWAHB
|
sosondowah ankyrin repeat domain family member B |
| chr7_+_106809406 | 0.18 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr15_+_59279851 | 0.18 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr4_-_76598296 | 0.18 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chrY_+_15016725 | 0.17 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr9_-_6007787 | 0.17 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr7_-_95064264 | 0.17 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr1_+_204494618 | 0.17 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr14_-_57735528 | 0.17 |
ENST00000340918.7
ENST00000413566.2 |
EXOC5
|
exocyst complex component 5 |
| chr4_-_152147579 | 0.17 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr1_+_57110972 | 0.17 |
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr5_+_95997918 | 0.17 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr4_-_175205407 | 0.17 |
ENST00000393674.2
|
FBXO8
|
F-box protein 8 |
| chr20_+_47662805 | 0.17 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr10_-_118764862 | 0.16 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr10_-_98346801 | 0.16 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr10_+_111767720 | 0.16 |
ENST00000356080.4
ENST00000277900.8 |
ADD3
|
adducin 3 (gamma) |
| chr15_-_52861394 | 0.16 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr3_+_130569429 | 0.16 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_+_95256356 | 0.16 |
ENST00000371485.3
|
CEP55
|
centrosomal protein 55kDa |
| chr3_-_178790057 | 0.15 |
ENST00000311417.2
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr2_+_103236004 | 0.15 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr11_+_34073195 | 0.15 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr3_+_186501336 | 0.15 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr10_+_93683519 | 0.15 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr9_+_19408999 | 0.15 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr17_+_28705921 | 0.15 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr3_-_138553594 | 0.15 |
ENST00000477593.1
ENST00000483968.1 |
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr6_-_53213780 | 0.15 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr12_-_22697343 | 0.15 |
ENST00000446597.1
ENST00000536386.1 ENST00000396028.2 ENST00000545552.1 ENST00000544930.1 ENST00000333957.4 |
C2CD5
|
C2 calcium-dependent domain containing 5 |
| chr12_+_56473628 | 0.15 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr4_-_71705590 | 0.15 |
ENST00000254799.6
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr5_+_86564739 | 0.14 |
ENST00000456692.2
ENST00000512763.1 ENST00000506290.1 |
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr17_+_55162453 | 0.14 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chrX_+_21392873 | 0.14 |
ENST00000379510.3
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr5_+_96271141 | 0.14 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr2_-_26101374 | 0.14 |
ENST00000435504.4
|
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr15_+_50716576 | 0.14 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr11_+_58346584 | 0.14 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr20_+_19867150 | 0.13 |
ENST00000255006.6
|
RIN2
|
Ras and Rab interactor 2 |
| chr6_-_132272504 | 0.13 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr8_-_12612962 | 0.13 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr1_+_101361626 | 0.13 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr2_+_64681219 | 0.13 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr3_-_64211112 | 0.13 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr9_-_4741255 | 0.13 |
ENST00000381809.3
|
AK3
|
adenylate kinase 3 |
| chrX_+_16804544 | 0.13 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr8_-_91658303 | 0.13 |
ENST00000458549.2
|
TMEM64
|
transmembrane protein 64 |
| chr14_-_64010046 | 0.13 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr2_+_173940442 | 0.13 |
ENST00000409176.2
ENST00000338983.3 ENST00000431503.2 |
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr12_+_12764773 | 0.13 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr1_+_109632425 | 0.12 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr3_+_113666748 | 0.12 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr10_+_89622870 | 0.12 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr19_+_41768401 | 0.12 |
ENST00000352456.3
ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr7_-_11871815 | 0.12 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr17_-_62658186 | 0.12 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr3_+_178866199 | 0.12 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr9_-_26947453 | 0.12 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr10_+_63661053 | 0.12 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr2_+_27070964 | 0.12 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr17_+_65821780 | 0.12 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr7_+_35840542 | 0.12 |
ENST00000435235.1
ENST00000399034.2 ENST00000350320.6 ENST00000469679.2 |
SEPT7
|
septin 7 |
| chr1_-_204380919 | 0.12 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr1_-_222885770 | 0.12 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr12_+_69004619 | 0.12 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr2_-_204400113 | 0.12 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr17_-_5372271 | 0.11 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chrX_-_131352152 | 0.11 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr6_-_111136513 | 0.11 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr13_-_27745936 | 0.11 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr12_-_50419177 | 0.11 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr4_+_85504075 | 0.11 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr2_-_122042770 | 0.11 |
ENST00000263707.5
|
TFCP2L1
|
transcription factor CP2-like 1 |
| chrX_+_41192595 | 0.11 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr17_+_30264014 | 0.11 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr10_+_23728198 | 0.11 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr2_+_30670077 | 0.11 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr16_+_19125252 | 0.11 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chrX_-_3631635 | 0.10 |
ENST00000262848.5
|
PRKX
|
protein kinase, X-linked |
| chr14_-_50698276 | 0.10 |
ENST00000216373.5
|
SOS2
|
son of sevenless homolog 2 (Drosophila) |
| chr8_-_105601134 | 0.10 |
ENST00000276654.5
ENST00000424843.2 |
LRP12
|
low density lipoprotein receptor-related protein 12 |
| chr5_+_138940742 | 0.10 |
ENST00000398733.3
ENST00000253815.2 ENST00000505007.1 |
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr3_-_151176497 | 0.10 |
ENST00000282466.3
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr1_+_93913713 | 0.10 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr17_+_57697216 | 0.10 |
ENST00000393043.1
ENST00000269122.3 |
CLTC
|
clathrin, heavy chain (Hc) |
| chr2_+_20646824 | 0.10 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr6_-_109703663 | 0.10 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chr2_+_173292301 | 0.10 |
ENST00000264106.6
ENST00000375221.2 ENST00000343713.4 |
ITGA6
|
integrin, alpha 6 |
| chr6_-_79787902 | 0.10 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr5_-_90679145 | 0.10 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr12_-_57082060 | 0.10 |
ENST00000448157.2
ENST00000414274.3 ENST00000262033.6 ENST00000456859.2 |
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr16_-_46723066 | 0.10 |
ENST00000299138.7
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr7_-_28220354 | 0.10 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr5_+_98104978 | 0.10 |
ENST00000308234.7
|
RGMB
|
repulsive guidance molecule family member b |
| chr1_+_28099683 | 0.10 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chrX_-_74376108 | 0.09 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr15_+_32907691 | 0.09 |
ENST00000361627.3
ENST00000567348.1 ENST00000563864.1 ENST00000543522.1 |
ARHGAP11A
|
Rho GTPase activating protein 11A |
| chr2_-_85555385 | 0.09 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr1_+_212458834 | 0.09 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr4_-_78740511 | 0.09 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr10_-_75173785 | 0.09 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chr8_-_22550815 | 0.09 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr16_-_46655538 | 0.09 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr3_+_152017181 | 0.09 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr9_-_111882195 | 0.09 |
ENST00000374586.3
|
TMEM245
|
transmembrane protein 245 |
| chr18_+_9913977 | 0.09 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr14_-_31495569 | 0.09 |
ENST00000357479.5
ENST00000355683.5 |
STRN3
|
striatin, calmodulin binding protein 3 |
| chr1_+_82266053 | 0.09 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr11_-_12030629 | 0.09 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr11_+_120207787 | 0.09 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_155829286 | 0.09 |
ENST00000368324.4
|
SYT11
|
synaptotagmin XI |
| chr7_-_27183263 | 0.09 |
ENST00000222726.3
|
HOXA5
|
homeobox A5 |
| chr6_+_25279651 | 0.09 |
ENST00000329474.6
|
LRRC16A
|
leucine rich repeat containing 16A |
| chr10_+_180987 | 0.09 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr13_-_30424821 | 0.09 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr7_-_105029329 | 0.09 |
ENST00000393651.3
ENST00000460391.1 |
SRPK2
|
SRSF protein kinase 2 |
| chr18_+_20513782 | 0.08 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr20_-_5591626 | 0.08 |
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr4_+_153701081 | 0.08 |
ENST00000451320.2
ENST00000429148.2 ENST00000353617.2 ENST00000405727.2 ENST00000356064.3 |
ARFIP1
|
ADP-ribosylation factor interacting protein 1 |
| chr2_+_118846008 | 0.08 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr1_+_42846443 | 0.08 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chrX_+_135067576 | 0.08 |
ENST00000370701.1
ENST00000370698.3 ENST00000370695.4 |
SLC9A6
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
| chr12_-_100536608 | 0.08 |
ENST00000356828.3
ENST00000279907.7 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr6_+_117996621 | 0.08 |
ENST00000368494.3
|
NUS1
|
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) |
| chr8_-_68255912 | 0.08 |
ENST00000262215.3
ENST00000519436.1 |
ARFGEF1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr16_+_21169976 | 0.08 |
ENST00000572258.1
ENST00000261388.3 ENST00000451578.2 ENST00000572599.1 ENST00000577162.1 |
TMEM159
|
transmembrane protein 159 |
| chr6_+_121756809 | 0.08 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr17_+_57784826 | 0.08 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr21_-_40685477 | 0.08 |
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr3_+_179370517 | 0.08 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr5_-_132073210 | 0.08 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr9_-_80646374 | 0.08 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr15_+_80987617 | 0.08 |
ENST00000258884.4
ENST00000558464.1 |
ABHD17C
|
abhydrolase domain containing 17C |
| chr2_-_182545603 | 0.08 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr14_+_96829814 | 0.08 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr13_+_80055284 | 0.08 |
ENST00000218652.7
|
NDFIP2
|
Nedd4 family interacting protein 2 |
| chr12_+_64845660 | 0.08 |
ENST00000331710.5
|
TBK1
|
TANK-binding kinase 1 |
| chr5_+_140345820 | 0.08 |
ENST00000289269.5
|
PCDHAC2
|
protocadherin alpha subfamily C, 2 |
| chr6_+_64281906 | 0.08 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_-_243418344 | 0.08 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr11_+_120894781 | 0.08 |
ENST00000529397.1
ENST00000528512.1 ENST00000422003.2 |
TBCEL
|
tubulin folding cofactor E-like |
| chr7_-_112579869 | 0.07 |
ENST00000297145.4
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr5_+_135468516 | 0.07 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr3_+_137906109 | 0.07 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr18_-_43547223 | 0.07 |
ENST00000282041.5
|
EPG5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr1_-_169337176 | 0.07 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chrX_-_34675391 | 0.07 |
ENST00000275954.3
|
TMEM47
|
transmembrane protein 47 |
| chr3_-_44519131 | 0.07 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr12_-_92539614 | 0.07 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_-_111846435 | 0.07 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_-_17104356 | 0.07 |
ENST00000361272.4
ENST00000523917.1 |
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr1_+_179923873 | 0.07 |
ENST00000367607.3
ENST00000491495.2 |
CEP350
|
centrosomal protein 350kDa |
| chr12_-_122907091 | 0.07 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr15_-_51914996 | 0.07 |
ENST00000251076.5
|
DMXL2
|
Dmx-like 2 |
| chr2_-_183903133 | 0.07 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr1_+_92495528 | 0.07 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr20_-_48330377 | 0.07 |
ENST00000371711.4
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr12_+_56511943 | 0.07 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr6_+_136172820 | 0.07 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr4_-_100867864 | 0.07 |
ENST00000442697.2
|
DNAJB14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr18_-_5296001 | 0.07 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr6_-_108279369 | 0.07 |
ENST00000369002.4
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr4_-_25864581 | 0.07 |
ENST00000399878.3
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_+_107879459 | 0.07 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr6_+_89790490 | 0.07 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr4_+_141445311 | 0.07 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr9_-_123964114 | 0.07 |
ENST00000373840.4
|
RAB14
|
RAB14, member RAS oncogene family |
| chr12_-_57824739 | 0.07 |
ENST00000347140.3
ENST00000402412.1 |
R3HDM2
|
R3H domain containing 2 |
| chr3_+_124223586 | 0.07 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr9_+_2015335 | 0.07 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_117186946 | 0.07 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr2_-_65357225 | 0.07 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr11_-_118661828 | 0.07 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr1_+_87380299 | 0.07 |
ENST00000370551.4
ENST00000370550.5 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr2_-_201828356 | 0.07 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
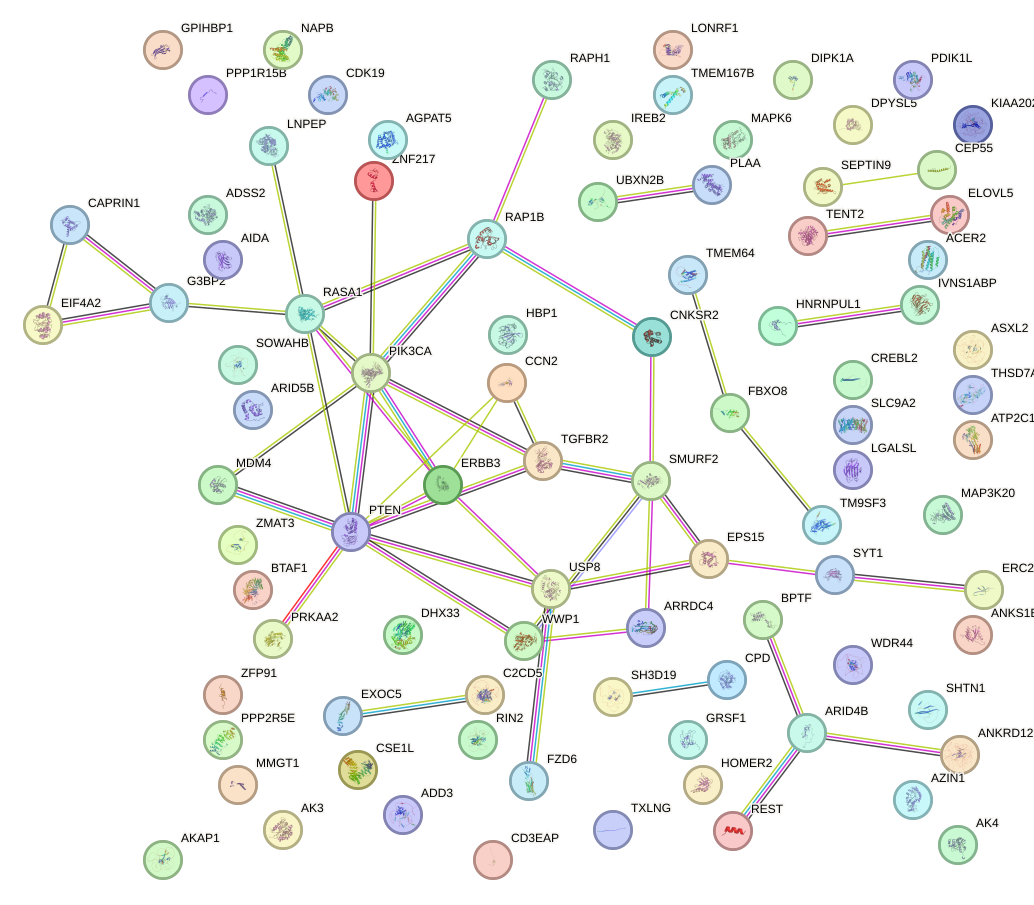
Network of associatons between targets according to the STRING database.
First level regulatory network of GUGCAAA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.2 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.1 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:1903179 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:1903121 | negative regulation of ribosome biogenesis(GO:0090071) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.2 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0044828 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.0 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.0 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.0 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.0 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.0 | 0.2 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.3 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) omegasome membrane(GO:1903349) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.2 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 0.2 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.1 | 0.2 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |