Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
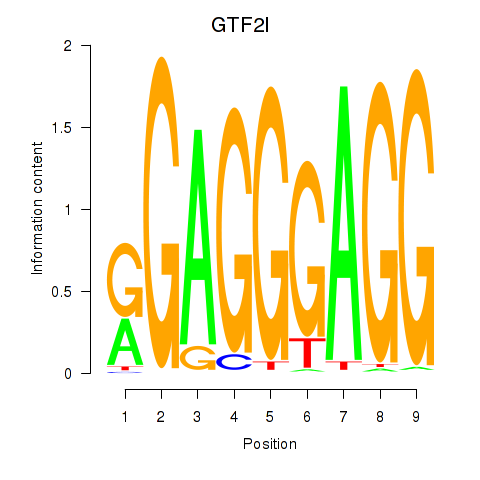
Results for GTF2I
Z-value: 1.38
Transcription factors associated with GTF2I
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GTF2I
|
ENSG00000077809.8 | general transcription factor IIi |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GTF2I | hg19_v2_chr7_+_74072288_74072357 | 0.86 | 1.4e-01 | Click! |
Activity profile of GTF2I motif
Sorted Z-values of GTF2I motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_81046841 | 0.98 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr2_+_219264762 | 0.92 |
ENST00000452977.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chrX_+_123095546 | 0.79 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr2_+_128175997 | 0.66 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr20_+_36531499 | 0.65 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr4_+_174089904 | 0.61 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chrX_+_123095155 | 0.60 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr6_-_10413112 | 0.58 |
ENST00000465858.1
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr12_+_123849462 | 0.58 |
ENST00000543072.1
|
hsa-mir-8072
|
hsa-mir-8072 |
| chr17_+_46131912 | 0.57 |
ENST00000584634.1
ENST00000580050.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr16_+_29817841 | 0.55 |
ENST00000322945.6
ENST00000562337.1 ENST00000566906.2 ENST00000563402.1 ENST00000219782.6 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr8_+_97506033 | 0.55 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr18_+_46065393 | 0.54 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr15_-_37392703 | 0.53 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr2_+_219264466 | 0.52 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr17_-_74533963 | 0.52 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr5_+_92919043 | 0.48 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr5_+_71403280 | 0.47 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr17_-_74533734 | 0.46 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr7_-_102252589 | 0.46 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr12_-_56881874 | 0.46 |
ENST00000539272.1
|
GLS2
|
glutaminase 2 (liver, mitochondrial) |
| chr2_-_174828892 | 0.45 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chr15_-_37392086 | 0.44 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr17_-_63052929 | 0.43 |
ENST00000439174.2
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13 |
| chr19_-_36297632 | 0.43 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr19_-_49567124 | 0.42 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr1_+_203444887 | 0.40 |
ENST00000343110.2
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein |
| chr10_-_88729200 | 0.40 |
ENST00000474994.2
|
MMRN2
|
multimerin 2 |
| chr5_+_176514413 | 0.40 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr3_-_123168551 | 0.39 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr1_-_235491462 | 0.39 |
ENST00000418304.1
ENST00000264183.3 ENST00000349213.3 |
ARID4B
|
AT rich interactive domain 4B (RBP1-like) |
| chr6_+_44095263 | 0.39 |
ENST00000532634.1
|
TMEM63B
|
transmembrane protein 63B |
| chr4_+_174089951 | 0.39 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr15_-_88799948 | 0.38 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr5_+_175298573 | 0.38 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr6_+_149638876 | 0.38 |
ENST00000392282.1
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr12_+_6930964 | 0.38 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr2_-_198364552 | 0.37 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr12_+_46123448 | 0.37 |
ENST00000334344.6
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr2_-_227664474 | 0.37 |
ENST00000305123.5
|
IRS1
|
insulin receptor substrate 1 |
| chr8_-_125486755 | 0.37 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr16_+_29817399 | 0.36 |
ENST00000545521.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr14_-_61191049 | 0.36 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr2_-_152684977 | 0.35 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr1_-_167906020 | 0.35 |
ENST00000458574.1
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr8_+_125486939 | 0.35 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr2_-_198364581 | 0.35 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr5_+_139027877 | 0.35 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chrX_-_77041685 | 0.35 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr7_-_21985489 | 0.35 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr14_-_61190754 | 0.34 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr4_-_105416039 | 0.34 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr17_-_7120498 | 0.33 |
ENST00000485100.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr17_-_46692457 | 0.33 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr3_+_20081515 | 0.33 |
ENST00000263754.4
|
KAT2B
|
K(lysine) acetyltransferase 2B |
| chr6_-_151712673 | 0.33 |
ENST00000325144.4
|
ZBTB2
|
zinc finger and BTB domain containing 2 |
| chr17_+_46131843 | 0.32 |
ENST00000577411.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr15_-_37393406 | 0.32 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr8_-_103136481 | 0.32 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chr19_+_45973120 | 0.32 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr4_-_83931862 | 0.32 |
ENST00000506560.1
ENST00000442461.2 ENST00000446851.2 ENST00000340417.3 |
LIN54
|
lin-54 homolog (C. elegans) |
| chr10_+_88718314 | 0.32 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr5_+_137673945 | 0.32 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr4_-_114682936 | 0.31 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chrX_+_123095860 | 0.31 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr9_-_35754253 | 0.31 |
ENST00000436428.2
|
MSMP
|
microseminoprotein, prostate associated |
| chrX_-_20284958 | 0.31 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr3_+_107244229 | 0.31 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr20_+_36531544 | 0.31 |
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr5_+_127419449 | 0.31 |
ENST00000262461.2
ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr1_+_167905894 | 0.31 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr5_+_176513895 | 0.30 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr12_-_124873357 | 0.30 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr6_+_33378517 | 0.30 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr12_-_56615485 | 0.30 |
ENST00000549038.1
ENST00000552244.1 |
RNF41
|
ring finger protein 41 |
| chr5_-_10308125 | 0.30 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr5_+_138089100 | 0.30 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr3_+_107318157 | 0.30 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr19_-_36297348 | 0.30 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr17_-_36891830 | 0.29 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr6_+_41514078 | 0.29 |
ENST00000373063.3
ENST00000373060.1 |
FOXP4
|
forkhead box P4 |
| chr9_-_34523027 | 0.29 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr7_-_21985656 | 0.29 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chrX_+_123095890 | 0.29 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr6_+_34482630 | 0.29 |
ENST00000538621.1
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr1_-_150946911 | 0.29 |
ENST00000457392.1
ENST00000421609.1 |
CERS2
|
ceramide synthase 2 |
| chr12_-_104531785 | 0.29 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr11_+_64950801 | 0.29 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr5_+_175298487 | 0.28 |
ENST00000393745.3
|
CPLX2
|
complexin 2 |
| chr16_+_29819372 | 0.28 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chrX_+_117629766 | 0.28 |
ENST00000276204.6
ENST00000276202.7 |
DOCK11
|
dedicator of cytokinesis 11 |
| chr12_+_6930813 | 0.28 |
ENST00000428545.2
|
GPR162
|
G protein-coupled receptor 162 |
| chr13_+_93879085 | 0.28 |
ENST00000377047.4
|
GPC6
|
glypican 6 |
| chr1_+_53793885 | 0.28 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr9_-_123476612 | 0.28 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr12_-_53893227 | 0.27 |
ENST00000547488.1
|
MAP3K12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr14_+_32546274 | 0.27 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr5_-_159739483 | 0.27 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr5_+_176513868 | 0.27 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr17_-_57232525 | 0.27 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_77179487 | 0.27 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr13_-_67804445 | 0.27 |
ENST00000456367.1
ENST00000377861.3 ENST00000544246.1 |
PCDH9
|
protocadherin 9 |
| chr2_+_42275153 | 0.27 |
ENST00000294964.5
|
PKDCC
|
protein kinase domain containing, cytoplasmic |
| chr2_+_189156638 | 0.26 |
ENST00000410051.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr12_+_56473628 | 0.26 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr9_-_123476719 | 0.26 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr12_-_6484376 | 0.26 |
ENST00000360168.3
ENST00000358945.3 |
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chr3_-_131753830 | 0.26 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr4_-_16228083 | 0.26 |
ENST00000399920.3
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr18_+_46065483 | 0.26 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr7_+_76139741 | 0.26 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr4_-_140477928 | 0.26 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr17_-_73839792 | 0.26 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_+_38219063 | 0.26 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chr5_-_142784003 | 0.26 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_+_6930703 | 0.25 |
ENST00000311268.3
|
GPR162
|
G protein-coupled receptor 162 |
| chrX_+_109245863 | 0.25 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr8_-_71520513 | 0.25 |
ENST00000262213.2
ENST00000536748.1 ENST00000518678.1 |
TRAM1
|
translocation associated membrane protein 1 |
| chr14_+_32546485 | 0.25 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_36024107 | 0.25 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr1_-_154193009 | 0.25 |
ENST00000368518.1
ENST00000368519.1 ENST00000368521.5 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr7_+_91570240 | 0.25 |
ENST00000394564.1
|
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chrX_-_71351678 | 0.24 |
ENST00000609883.1
ENST00000545866.1 |
RGAG4
|
retrotransposon gag domain containing 4 |
| chr1_-_150947299 | 0.24 |
ENST00000361419.5
|
CERS2
|
ceramide synthase 2 |
| chr17_-_15165825 | 0.24 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr3_-_52486841 | 0.24 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr1_-_9970227 | 0.24 |
ENST00000377263.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr6_-_86353510 | 0.24 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr5_+_139028510 | 0.24 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr14_-_102605983 | 0.24 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr20_-_49547910 | 0.24 |
ENST00000396032.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr12_-_123849374 | 0.24 |
ENST00000602398.1
ENST00000602750.1 |
SBNO1
|
strawberry notch homolog 1 (Drosophila) |
| chr12_-_48213568 | 0.23 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr19_+_46367518 | 0.23 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr1_-_245026388 | 0.23 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr5_+_95998070 | 0.23 |
ENST00000421689.2
ENST00000510756.1 ENST00000512620.1 |
CAST
|
calpastatin |
| chr17_+_46132037 | 0.23 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr7_+_65552756 | 0.23 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr4_-_76598296 | 0.23 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr12_-_53625958 | 0.23 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr4_+_129731074 | 0.23 |
ENST00000512960.1
ENST00000503785.1 ENST00000514740.1 |
PHF17
|
jade family PHD finger 1 |
| chr12_-_48213735 | 0.23 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr20_-_62715712 | 0.23 |
ENST00000308906.2
ENST00000302096.4 |
C20orf201
|
chromosome 20 open reading frame 201 |
| chr16_+_29819446 | 0.23 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr1_-_151299842 | 0.23 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr7_+_76139925 | 0.23 |
ENST00000394849.1
|
UPK3B
|
uroplakin 3B |
| chr7_+_100797726 | 0.23 |
ENST00000429457.1
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr10_-_88729069 | 0.22 |
ENST00000609457.1
|
MMRN2
|
multimerin 2 |
| chr7_+_87563557 | 0.22 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr20_-_49547731 | 0.22 |
ENST00000396029.3
|
ADNP
|
activity-dependent neuroprotector homeobox |
| chr17_+_48243352 | 0.22 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr16_-_66730583 | 0.22 |
ENST00000330687.4
ENST00000394106.2 ENST00000563952.1 |
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chrX_-_70474910 | 0.22 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr7_-_102252038 | 0.22 |
ENST00000461209.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr16_-_57513657 | 0.22 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr19_-_49149553 | 0.22 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr15_+_93443419 | 0.22 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_101733794 | 0.22 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_+_95997885 | 0.22 |
ENST00000511097.2
|
CAST
|
calpastatin |
| chr16_+_57662419 | 0.22 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr12_+_94542459 | 0.22 |
ENST00000258526.4
|
PLXNC1
|
plexin C1 |
| chr12_+_41086297 | 0.22 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr1_-_167906277 | 0.22 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr7_-_100425112 | 0.22 |
ENST00000358173.3
|
EPHB4
|
EPH receptor B4 |
| chr1_+_109102652 | 0.22 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr10_-_103578182 | 0.22 |
ENST00000439817.1
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr19_-_36499521 | 0.21 |
ENST00000397428.3
ENST00000503121.1 ENST00000340477.5 ENST00000324444.3 ENST00000490730.1 |
SYNE4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr16_+_57662527 | 0.21 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_-_52580920 | 0.21 |
ENST00000219746.9
|
TOX3
|
TOX high mobility group box family member 3 |
| chr2_-_19558373 | 0.21 |
ENST00000272223.2
|
OSR1
|
odd-skipped related transciption factor 1 |
| chr2_-_75788428 | 0.21 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr10_+_104503727 | 0.21 |
ENST00000448841.1
|
WBP1L
|
WW domain binding protein 1-like |
| chr19_-_50990785 | 0.21 |
ENST00000595005.1
|
CTD-2545M3.8
|
CTD-2545M3.8 |
| chr1_+_26438289 | 0.21 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr2_-_45165994 | 0.21 |
ENST00000444871.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr12_+_57853918 | 0.21 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr1_+_211433275 | 0.21 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr5_+_95997918 | 0.21 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr11_+_18344106 | 0.21 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr12_-_89918522 | 0.21 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr19_-_39368887 | 0.21 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr17_+_38278530 | 0.21 |
ENST00000398532.4
|
MSL1
|
male-specific lethal 1 homolog (Drosophila) |
| chr12_-_104532062 | 0.21 |
ENST00000240055.3
|
NFYB
|
nuclear transcription factor Y, beta |
| chr8_+_27491381 | 0.21 |
ENST00000337221.4
|
SCARA3
|
scavenger receptor class A, member 3 |
| chr1_+_167906056 | 0.20 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr4_+_129730839 | 0.20 |
ENST00000511647.1
|
PHF17
|
jade family PHD finger 1 |
| chr5_-_137674000 | 0.20 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr19_+_45973360 | 0.20 |
ENST00000589593.1
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr17_-_73840415 | 0.20 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr12_+_107168418 | 0.20 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr12_-_80328700 | 0.20 |
ENST00000550107.1
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A |
| chr3_-_33481835 | 0.20 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chrX_-_107019181 | 0.20 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_+_41860047 | 0.20 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr6_+_41514305 | 0.19 |
ENST00000409208.1
ENST00000373057.3 |
FOXP4
|
forkhead box P4 |
| chr6_+_34433844 | 0.19 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr5_+_175223313 | 0.19 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr22_-_27620603 | 0.19 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr12_+_53845879 | 0.19 |
ENST00000359282.5
ENST00000603815.1 ENST00000447282.1 ENST00000437231.1 ENST00000549863.1 ENST00000359462.5 ENST00000550520.2 ENST00000546463.1 ENST00000552296.2 |
PCBP2
|
poly(rC) binding protein 2 |
| chr17_-_47755436 | 0.19 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of GTF2I
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.7 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.7 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.2 | 0.2 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.2 | 1.0 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 0.6 | GO:0014011 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.1 | 0.6 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.6 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 1.0 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.4 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 1.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.7 | GO:0021622 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.5 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.1 | 0.7 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.3 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.3 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 0.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.4 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.5 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.2 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.1 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.1 | 0.5 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.2 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.1 | 0.2 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.3 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.2 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.7 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 0.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.1 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.5 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.8 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0035811 | negative regulation of urine volume(GO:0035811) response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:1903382 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.8 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.3 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.7 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) |
| 0.0 | 0.8 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.6 | GO:2000675 | regulation of type B pancreatic cell apoptotic process(GO:2000674) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 1.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) |
| 0.0 | 0.3 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:1901804 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.2 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.0 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.5 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.3 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.1 | GO:0048298 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.0 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 1.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0010693 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) platelet dense granule(GO:0042827) |
| 0.1 | 0.3 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.2 | 0.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.4 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.3 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.2 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.1 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.3 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.2 | GO:0016730 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.0 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0052840 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 2.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |