Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
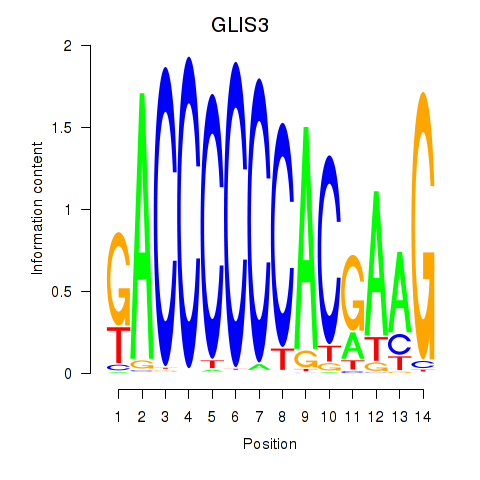
Results for GLIS3
Z-value: 0.25
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLIS3 | hg19_v2_chr9_-_4299874_4299916 | -0.47 | 5.3e-01 | Click! |
Activity profile of GLIS3 motif
Sorted Z-values of GLIS3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_4675175 | 0.32 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr11_+_68451943 | 0.21 |
ENST00000265643.3
|
GAL
|
galanin/GMAP prepropeptide |
| chr1_+_16062820 | 0.16 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr3_-_160167540 | 0.15 |
ENST00000496222.1
ENST00000471396.1 ENST00000471155.1 ENST00000309784.4 |
TRIM59
|
tripartite motif containing 59 |
| chr5_+_172386517 | 0.13 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26-like 1 |
| chr5_+_41925325 | 0.10 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr18_+_11752040 | 0.09 |
ENST00000423027.3
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr13_-_20767037 | 0.09 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr19_+_1495362 | 0.09 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr19_+_41281060 | 0.09 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr5_+_72509751 | 0.08 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr14_+_105190514 | 0.08 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr9_-_139658965 | 0.08 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr16_+_89686991 | 0.08 |
ENST00000393092.3
|
DPEP1
|
dipeptidase 1 (renal) |
| chr7_+_65958693 | 0.08 |
ENST00000445681.1
ENST00000452565.1 |
GS1-124K5.4
|
GS1-124K5.4 |
| chr5_-_137090028 | 0.08 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr1_-_53018654 | 0.08 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr5_+_172386419 | 0.07 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr1_-_22215192 | 0.07 |
ENST00000374673.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr1_-_247095236 | 0.07 |
ENST00000478568.1
|
AHCTF1
|
AT hook containing transcription factor 1 |
| chr17_-_5015129 | 0.06 |
ENST00000575898.1
ENST00000416429.2 |
ZNF232
|
zinc finger protein 232 |
| chr11_+_393428 | 0.06 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr8_-_29120604 | 0.06 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr7_+_150758642 | 0.06 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr22_+_18834324 | 0.05 |
ENST00000342005.4
|
AC008132.13
|
Uncharacterized protein |
| chr17_-_33446820 | 0.05 |
ENST00000592577.1
ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D
|
RAD51 paralog D |
| chr17_-_73178599 | 0.05 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr17_-_73179046 | 0.05 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr20_+_18447771 | 0.05 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr9_-_98279241 | 0.04 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr4_+_164265035 | 0.04 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr3_-_25824872 | 0.04 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr1_-_53019059 | 0.04 |
ENST00000484723.2
ENST00000524582.1 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr19_-_48673465 | 0.04 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr18_+_11751493 | 0.04 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr8_-_74791051 | 0.04 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr17_+_80818231 | 0.04 |
ENST00000576996.1
|
TBCD
|
tubulin folding cofactor D |
| chr11_+_66886717 | 0.04 |
ENST00000398645.2
|
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr7_-_50518022 | 0.04 |
ENST00000356889.4
ENST00000420829.1 ENST00000448788.1 ENST00000395556.2 ENST00000422854.1 ENST00000435566.1 ENST00000433017.1 |
FIGNL1
|
fidgetin-like 1 |
| chr6_+_89791507 | 0.04 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr1_+_110527308 | 0.04 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr17_-_3819751 | 0.03 |
ENST00000225538.3
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr3_+_127348005 | 0.03 |
ENST00000342480.6
|
PODXL2
|
podocalyxin-like 2 |
| chr6_+_44191290 | 0.03 |
ENST00000371755.3
ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr1_-_157108266 | 0.03 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr6_+_44191507 | 0.03 |
ENST00000371724.1
ENST00000371713.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr1_+_59250815 | 0.03 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr1_-_42800860 | 0.03 |
ENST00000445886.1
ENST00000361346.1 ENST00000361776.1 |
FOXJ3
|
forkhead box J3 |
| chr11_+_134855246 | 0.03 |
ENST00000597621.1
|
AP003062.1
|
CDNA FLJ27342 fis, clone TST02993; Uncharacterized protein |
| chr17_+_7792101 | 0.03 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr6_+_24495067 | 0.03 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr8_-_12051576 | 0.02 |
ENST00000524571.2
ENST00000533852.2 ENST00000533513.1 ENST00000448228.2 ENST00000534520.1 ENST00000321602.8 |
FAM86B1
|
family with sequence similarity 86, member B1 |
| chr20_-_44718538 | 0.02 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr11_-_68671264 | 0.02 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr1_-_42801540 | 0.02 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr19_-_14224969 | 0.02 |
ENST00000589994.1
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chrX_+_118370288 | 0.02 |
ENST00000535419.1
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr20_-_23969416 | 0.02 |
ENST00000335694.4
|
GGTLC1
|
gamma-glutamyltransferase light chain 1 |
| chr3_-_160167508 | 0.02 |
ENST00000479460.1
|
TRIM59
|
tripartite motif containing 59 |
| chr11_+_68671310 | 0.02 |
ENST00000255078.3
ENST00000539224.1 |
IGHMBP2
|
immunoglobulin mu binding protein 2 |
| chr9_-_130742792 | 0.01 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr1_+_33207381 | 0.01 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr7_+_23636992 | 0.01 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr15_+_91478493 | 0.01 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr4_-_84255935 | 0.01 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr9_-_88874519 | 0.01 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr19_-_56092187 | 0.01 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chrX_+_64887512 | 0.01 |
ENST00000360270.5
|
MSN
|
moesin |
| chr17_-_77925806 | 0.01 |
ENST00000574241.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr6_+_24495185 | 0.01 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr22_+_50628999 | 0.01 |
ENST00000395827.1
|
TRABD
|
TraB domain containing |
| chr6_+_33172407 | 0.01 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr17_+_5389605 | 0.01 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr16_-_3285144 | 0.01 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr20_-_18447667 | 0.01 |
ENST00000262547.5
ENST00000329494.5 ENST00000357236.4 |
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr11_-_18813110 | 0.01 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr18_+_3451584 | 0.00 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_-_25824925 | 0.00 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr9_-_116061476 | 0.00 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr11_-_8680383 | 0.00 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr2_-_241759622 | 0.00 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr1_+_1950763 | 0.00 |
ENST00000378585.4
|
GABRD
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr2_-_241737128 | 0.00 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr21_-_46330545 | 0.00 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLIS3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0043605 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.0 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0008283 | cell proliferation(GO:0008283) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.0 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |