Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GLI2
Z-value: 0.76
Transcription factors associated with GLI2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI2
|
ENSG00000074047.16 | GLI family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI2 | hg19_v2_chr2_+_121493717_121493823 | 0.18 | 8.2e-01 | Click! |
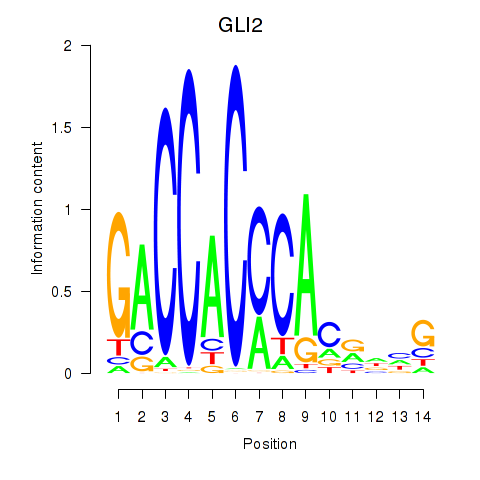
Activity profile of GLI2 motif
Sorted Z-values of GLI2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_76101379 | 0.33 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr19_+_44084696 | 0.31 |
ENST00000562255.1
ENST00000569031.2 |
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr3_+_184279566 | 0.31 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chr21_-_35340759 | 0.29 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr22_-_21482352 | 0.28 |
ENST00000329949.3
|
POM121L7
|
POM121 transmembrane nucleoporin-like 7 |
| chr3_+_112930387 | 0.27 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr22_+_20008646 | 0.26 |
ENST00000401833.1
|
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr15_+_29211570 | 0.25 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr8_+_7705398 | 0.25 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr19_-_36288740 | 0.24 |
ENST00000564335.1
ENST00000567313.1 ENST00000433059.1 |
AC002398.5
|
AC002398.5 |
| chr8_-_7320974 | 0.24 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr22_-_36761081 | 0.24 |
ENST00000456729.1
ENST00000401701.1 |
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr16_+_68279256 | 0.24 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr17_-_26989136 | 0.23 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr19_-_17571722 | 0.23 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr16_-_4664382 | 0.22 |
ENST00000591113.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr8_+_96146168 | 0.22 |
ENST00000519516.1
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr3_+_88108381 | 0.21 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr22_+_45714672 | 0.21 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr12_-_2944184 | 0.21 |
ENST00000337508.4
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr17_-_79917645 | 0.20 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr17_-_71258019 | 0.20 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr21_-_45079341 | 0.20 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr22_+_45714361 | 0.19 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr5_+_172332220 | 0.19 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr1_+_47799542 | 0.19 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr17_+_79071365 | 0.19 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr2_+_219264762 | 0.19 |
ENST00000452977.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr19_-_44084586 | 0.18 |
ENST00000599693.1
ENST00000598165.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr10_+_71389983 | 0.18 |
ENST00000373279.4
|
C10orf35
|
chromosome 10 open reading frame 35 |
| chr1_+_155051379 | 0.18 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr4_-_156298028 | 0.18 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr3_+_188817891 | 0.18 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr1_+_179263308 | 0.17 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr12_+_116985896 | 0.17 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr7_+_106810165 | 0.17 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr1_-_153433120 | 0.17 |
ENST00000368723.3
|
S100A7
|
S100 calcium binding protein A7 |
| chr7_-_92465868 | 0.17 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr2_+_237476419 | 0.17 |
ENST00000447924.1
|
ACKR3
|
atypical chemokine receptor 3 |
| chr17_-_30185946 | 0.17 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr20_-_25062767 | 0.17 |
ENST00000429762.3
ENST00000444511.2 ENST00000376707.3 |
VSX1
|
visual system homeobox 1 |
| chr7_-_151511911 | 0.16 |
ENST00000392801.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_+_152178320 | 0.16 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr1_-_2718286 | 0.16 |
ENST00000401094.6
|
TTC34
|
tetratricopeptide repeat domain 34 |
| chr3_-_10052849 | 0.16 |
ENST00000437616.1
ENST00000429065.2 |
AC022007.5
|
AC022007.5 |
| chr22_+_37447771 | 0.16 |
ENST00000402077.3
ENST00000403888.3 ENST00000456470.1 |
KCTD17
|
potassium channel tetramerization domain containing 17 |
| chr22_-_21482046 | 0.16 |
ENST00000419447.1
|
POM121L7
|
POM121 transmembrane nucleoporin-like 7 |
| chr15_+_40731920 | 0.16 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr12_+_69202975 | 0.16 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr19_+_56915668 | 0.15 |
ENST00000333201.9
ENST00000391778.3 |
ZNF583
|
zinc finger protein 583 |
| chr11_-_118927561 | 0.15 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr9_-_35071960 | 0.15 |
ENST00000417448.1
|
VCP
|
valosin containing protein |
| chr15_-_78933567 | 0.15 |
ENST00000261751.3
ENST00000412074.2 |
CHRNB4
|
cholinergic receptor, nicotinic, beta 4 (neuronal) |
| chr6_-_74233480 | 0.15 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_-_65321198 | 0.15 |
ENST00000530426.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr19_-_10613862 | 0.14 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr17_-_74582191 | 0.14 |
ENST00000225276.5
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr19_-_18197714 | 0.14 |
ENST00000322153.7
|
IL12RB1
|
interleukin 12 receptor, beta 1 |
| chr3_-_88108192 | 0.14 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr16_+_50059182 | 0.14 |
ENST00000562576.1
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr12_+_51633061 | 0.14 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr20_+_32782375 | 0.14 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr17_+_46184911 | 0.14 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr22_+_44427230 | 0.14 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr3_-_15140629 | 0.14 |
ENST00000507357.1
ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20
|
zinc finger, FYVE domain containing 20 |
| chr16_+_55522536 | 0.14 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr19_+_45165545 | 0.14 |
ENST00000592472.1
ENST00000587729.1 ENST00000585657.1 ENST00000592789.1 ENST00000591979.1 |
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr17_+_29248918 | 0.14 |
ENST00000581548.1
ENST00000580525.1 |
ADAP2
|
ArfGAP with dual PH domains 2 |
| chr15_-_75917955 | 0.13 |
ENST00000568162.1
ENST00000563875.1 |
SNUPN
|
snurportin 1 |
| chr17_+_46132037 | 0.13 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr16_+_2880157 | 0.13 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr3_+_10183447 | 0.13 |
ENST00000345392.2
|
VHL
|
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
| chr1_-_209792111 | 0.13 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chr1_+_161195781 | 0.13 |
ENST00000367988.3
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chr1_+_161195835 | 0.13 |
ENST00000545897.1
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
| chrX_+_23685653 | 0.13 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr6_-_26027480 | 0.12 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr3_+_32993065 | 0.12 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr19_+_17638059 | 0.12 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr5_+_154238149 | 0.12 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr22_+_20008595 | 0.12 |
ENST00000398042.2
ENST00000450664.1 ENST00000327374.4 ENST00000432883.1 |
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr5_+_131409476 | 0.12 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr10_+_104474353 | 0.12 |
ENST00000602647.1
ENST00000602439.1 ENST00000602764.1 |
SFXN2
|
sideroflexin 2 |
| chr8_+_94752349 | 0.12 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr11_-_63439013 | 0.12 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chrX_+_17755563 | 0.12 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr16_+_15744078 | 0.12 |
ENST00000396354.1
ENST00000570727.1 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr9_-_20382446 | 0.12 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr11_+_101918153 | 0.12 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr11_+_111957497 | 0.11 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr8_-_42065187 | 0.11 |
ENST00000270189.6
ENST00000352041.3 ENST00000220809.4 |
PLAT
|
plasminogen activator, tissue |
| chr19_-_6481776 | 0.11 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr20_-_13619550 | 0.11 |
ENST00000455532.1
ENST00000544472.1 ENST00000539805.1 ENST00000337743.4 |
TASP1
|
taspase, threonine aspartase, 1 |
| chr16_-_4817129 | 0.11 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr16_+_75252878 | 0.11 |
ENST00000361017.4
|
CTRB1
|
chymotrypsinogen B1 |
| chr10_+_104503727 | 0.11 |
ENST00000448841.1
|
WBP1L
|
WW domain binding protein 1-like |
| chr12_-_110883346 | 0.11 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr17_-_79881408 | 0.11 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr16_+_68279207 | 0.11 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr5_+_89770664 | 0.11 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr12_-_90024360 | 0.11 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr3_-_127317047 | 0.11 |
ENST00000462228.1
ENST00000490643.1 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr22_-_31688431 | 0.11 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr19_-_13947099 | 0.11 |
ENST00000587762.1
|
MIR24-2
|
microRNA 24-2 |
| chr5_-_89705537 | 0.11 |
ENST00000522864.1
ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3
|
centrin, EF-hand protein, 3 |
| chr15_+_75315896 | 0.10 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr22_+_20008678 | 0.10 |
ENST00000434168.1
|
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr11_-_71751715 | 0.10 |
ENST00000535947.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr17_+_74734052 | 0.10 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr10_-_120938303 | 0.10 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr3_+_160939050 | 0.10 |
ENST00000493066.1
ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr7_-_75452673 | 0.10 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr7_-_42971759 | 0.10 |
ENST00000538645.1
ENST00000445517.1 ENST00000223321.4 |
PSMA2
|
proteasome (prosome, macropain) subunit, alpha type, 2 |
| chr16_+_31885079 | 0.10 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr1_+_153600869 | 0.10 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr16_-_57832004 | 0.10 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr9_+_19408919 | 0.10 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr3_-_27410847 | 0.10 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr14_-_91976874 | 0.10 |
ENST00000557018.1
|
SMEK1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chrX_-_135056106 | 0.10 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr12_-_122711968 | 0.10 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr2_-_70475586 | 0.10 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_-_64527425 | 0.10 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr15_+_52311398 | 0.10 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr15_-_50838888 | 0.10 |
ENST00000532404.1
|
USP50
|
ubiquitin specific peptidase 50 |
| chr6_-_29648887 | 0.10 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr18_+_56531584 | 0.10 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr12_+_31227192 | 0.10 |
ENST00000535317.1
|
DDX11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr22_+_25465786 | 0.10 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr7_-_112430427 | 0.10 |
ENST00000449743.1
ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168
|
transmembrane protein 168 |
| chr12_+_110338323 | 0.10 |
ENST00000312777.5
ENST00000536408.2 |
TCHP
|
trichoplein, keratin filament binding |
| chr8_-_128960591 | 0.10 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
| chr1_-_32403370 | 0.09 |
ENST00000534796.1
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr19_+_19174795 | 0.09 |
ENST00000318596.7
|
SLC25A42
|
solute carrier family 25, member 42 |
| chr9_-_139658965 | 0.09 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr9_-_123476612 | 0.09 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr1_-_20834586 | 0.09 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr19_+_19144384 | 0.09 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr15_-_59225758 | 0.09 |
ENST00000558486.1
ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM
|
SAFB-like, transcription modulator |
| chr15_+_79603404 | 0.09 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chr3_-_56717246 | 0.09 |
ENST00000355628.5
|
FAM208A
|
family with sequence similarity 208, member A |
| chr19_-_37406923 | 0.09 |
ENST00000520965.1
|
ZNF829
|
zinc finger protein 829 |
| chr19_+_35810164 | 0.09 |
ENST00000598537.1
|
CD22
|
CD22 molecule |
| chr3_-_23958402 | 0.09 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr17_-_17723746 | 0.09 |
ENST00000577897.1
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr11_+_66025091 | 0.09 |
ENST00000526758.1
ENST00000440228.2 |
KLC2
|
kinesin light chain 2 |
| chr17_+_79213039 | 0.09 |
ENST00000431388.2
|
C17orf89
|
chromosome 17 open reading frame 89 |
| chr12_+_110338063 | 0.09 |
ENST00000405876.4
|
TCHP
|
trichoplein, keratin filament binding |
| chr3_-_185542761 | 0.09 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_+_17337406 | 0.09 |
ENST00000597836.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr11_-_12030629 | 0.09 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr10_-_101989315 | 0.09 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr13_+_113760098 | 0.09 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr22_+_51176624 | 0.09 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr22_-_24093267 | 0.09 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70 |
| chr16_-_84538218 | 0.09 |
ENST00000562447.1
ENST00000565765.1 ENST00000535580.1 ENST00000343629.6 |
TLDC1
|
TBC/LysM-associated domain containing 1 |
| chr7_-_43965937 | 0.09 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr4_+_72052964 | 0.09 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr3_+_127317066 | 0.09 |
ENST00000265056.7
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr13_-_60738003 | 0.09 |
ENST00000400330.1
ENST00000400324.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr17_+_42219267 | 0.09 |
ENST00000319977.4
ENST00000585683.1 |
C17orf53
|
chromosome 17 open reading frame 53 |
| chr14_+_96342729 | 0.09 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr17_+_80009330 | 0.09 |
ENST00000580716.1
ENST00000583961.1 |
GPS1
|
G protein pathway suppressor 1 |
| chr19_+_1908013 | 0.09 |
ENST00000454697.1
|
ADAT3
|
adenosine deaminase, tRNA-specific 3 |
| chr2_+_65215604 | 0.09 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr4_-_39529049 | 0.09 |
ENST00000501493.2
ENST00000509391.1 ENST00000507089.1 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr10_-_38265517 | 0.09 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr4_+_3344141 | 0.09 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr3_-_49449350 | 0.09 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr7_-_95064264 | 0.09 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr19_-_40791211 | 0.08 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr7_+_142985467 | 0.08 |
ENST00000392925.2
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr19_+_17638041 | 0.08 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr3_+_15469058 | 0.08 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chr1_+_179262905 | 0.08 |
ENST00000539888.1
ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1
|
sterol O-acyltransferase 1 |
| chr14_+_105559784 | 0.08 |
ENST00000548104.1
|
RP11-44N21.1
|
RP11-44N21.1 |
| chr8_-_145642203 | 0.08 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr16_+_57653854 | 0.08 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr21_-_46964306 | 0.08 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr19_+_35820064 | 0.08 |
ENST00000341773.6
ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22
|
CD22 molecule |
| chr1_+_47799446 | 0.08 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr12_+_57853918 | 0.08 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr16_-_11681316 | 0.08 |
ENST00000571688.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr16_+_58033450 | 0.08 |
ENST00000561743.1
|
USB1
|
U6 snRNA biogenesis 1 |
| chr22_+_29138013 | 0.08 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr11_+_20409227 | 0.08 |
ENST00000437750.2
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr2_+_11295624 | 0.08 |
ENST00000402361.1
ENST00000428481.1 |
PQLC3
|
PQ loop repeat containing 3 |
| chr1_-_36185073 | 0.08 |
ENST00000270815.4
|
C1orf216
|
chromosome 1 open reading frame 216 |
| chr1_+_3388181 | 0.08 |
ENST00000418137.1
ENST00000413250.2 |
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr2_+_26467825 | 0.08 |
ENST00000545822.1
ENST00000425035.1 |
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr2_-_96874553 | 0.08 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr15_-_102192567 | 0.08 |
ENST00000333202.3
ENST00000428002.2 ENST00000559107.1 ENST00000347970.3 |
TM2D3
|
TM2 domain containing 3 |
| chr18_-_77793891 | 0.08 |
ENST00000592957.1
ENST00000585474.1 |
TXNL4A
|
thioredoxin-like 4A |
| chr3_-_10052869 | 0.08 |
ENST00000454232.1
|
AC022007.5
|
AC022007.5 |
| chr14_+_100531738 | 0.08 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr10_+_75910960 | 0.08 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chr15_+_91478493 | 0.08 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr3_-_38691119 | 0.08 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr11_-_118789613 | 0.07 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr22_+_29168652 | 0.07 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.2 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.1 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.3 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.2 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0072387 | flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:1903381 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0015880 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.0 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.2 | GO:1904627 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) negative regulation of monocyte differentiation(GO:0045656) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.0 | GO:2000282 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:1902739 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.0 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.2 | GO:0097513 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:0031982 | vesicle(GO:0031982) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.0 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.0 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 0.0 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0015186 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |