Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GGAGUGU
Z-value: 0.31
miRNA associated with seed GGAGUGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-122-5p
|
MIMAT0000421 |
Activity profile of GGAGUGU motif
Sorted Z-values of GGAGUGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_162864575 | 0.36 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr1_+_28586006 | 0.17 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr4_-_76598296 | 0.17 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_-_95225768 | 0.14 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr7_+_94285637 | 0.11 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr1_+_16062820 | 0.10 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr4_-_53525406 | 0.10 |
ENST00000451218.2
ENST00000441222.3 |
USP46
|
ubiquitin specific peptidase 46 |
| chr1_-_244615425 | 0.08 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase |
| chr10_-_32345305 | 0.07 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr2_-_96811170 | 0.07 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr20_-_31071239 | 0.07 |
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr6_+_108881012 | 0.05 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr2_-_86790593 | 0.05 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr19_-_2456922 | 0.05 |
ENST00000582871.1
ENST00000325327.3 |
LMNB2
|
lamin B2 |
| chrX_-_135056216 | 0.05 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr2_-_86948245 | 0.05 |
ENST00000439940.2
ENST00000604011.1 |
CHMP3
RNF103-CHMP3
|
charged multivesicular body protein 3 RNF103-CHMP3 readthrough |
| chr4_+_172734548 | 0.05 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr10_+_70480963 | 0.04 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr14_+_37667118 | 0.04 |
ENST00000556615.1
ENST00000327441.7 ENST00000536774.1 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr19_+_46800289 | 0.04 |
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr3_+_9773409 | 0.04 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr19_-_47291843 | 0.04 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr11_-_108369101 | 0.04 |
ENST00000323468.5
|
KDELC2
|
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr8_-_117886955 | 0.03 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr17_-_27916621 | 0.03 |
ENST00000225394.3
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr8_-_68255912 | 0.03 |
ENST00000262215.3
ENST00000519436.1 |
ARFGEF1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr5_-_141392538 | 0.03 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_-_109940550 | 0.02 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr17_-_4269768 | 0.02 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr16_-_31076332 | 0.02 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr1_-_245027833 | 0.02 |
ENST00000444376.2
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr12_-_56694142 | 0.02 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr15_+_44719394 | 0.02 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr5_+_96271141 | 0.02 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr12_-_98897617 | 0.01 |
ENST00000501499.1
|
RP11-181C3.1
|
Uncharacterized protein |
| chr15_-_34628951 | 0.01 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr1_+_25071848 | 0.01 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr20_+_32581452 | 0.01 |
ENST00000375114.3
ENST00000448364.1 |
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr10_-_23003460 | 0.01 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr4_-_103266219 | 0.01 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr6_-_35464727 | 0.01 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr10_-_74856608 | 0.01 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr18_-_47013586 | 0.01 |
ENST00000318240.3
ENST00000579820.1 |
C18orf32
|
chromosome 18 open reading frame 32 |
| chr17_-_19880992 | 0.00 |
ENST00000395536.3
ENST00000576896.1 ENST00000225737.6 |
AKAP10
|
A kinase (PRKA) anchor protein 10 |
| chr15_-_83316254 | 0.00 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr1_-_70820357 | 0.00 |
ENST00000370944.4
ENST00000262346.6 |
ANKRD13C
|
ankyrin repeat domain 13C |
| chr17_+_41857793 | 0.00 |
ENST00000449302.3
|
C17orf105
|
chromosome 17 open reading frame 105 |
| chr10_-_61666267 | 0.00 |
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6 |
| chr16_-_30381580 | 0.00 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chrX_-_48901012 | 0.00 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
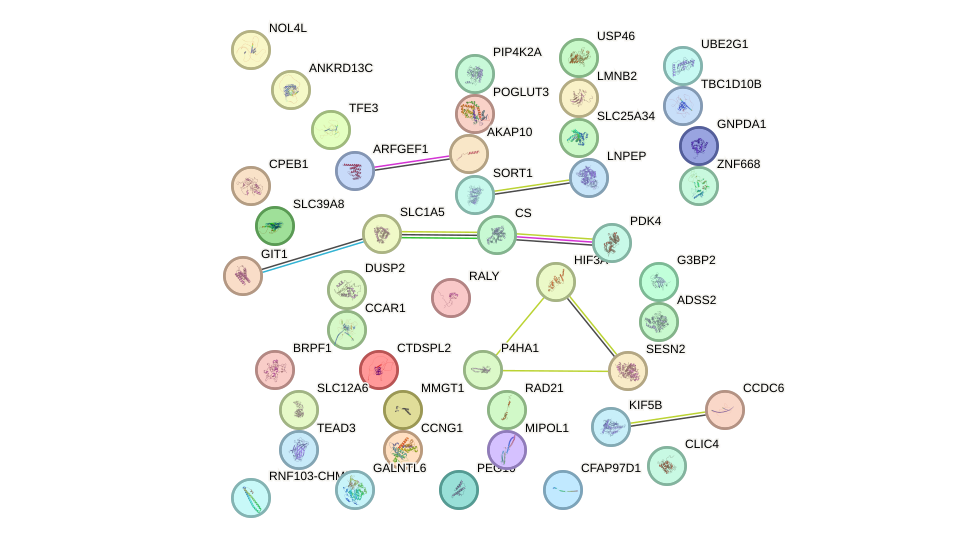
Network of associatons between targets according to the STRING database.
First level regulatory network of GGAGUGU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |