Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GFI1
Z-value: 0.54
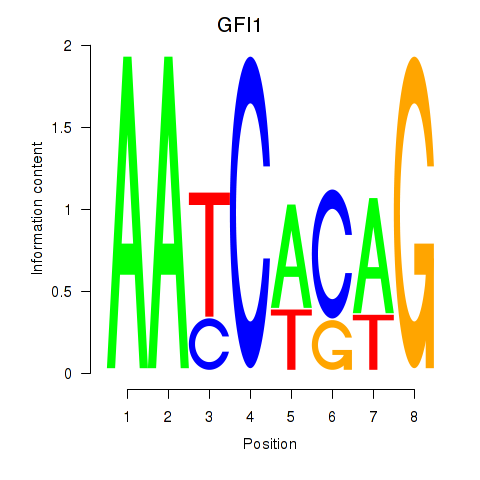
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | growth factor independent 1 transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92949505_92949543 | -0.53 | 4.7e-01 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_119056178 | 0.38 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr2_-_61389240 | 0.37 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr4_-_155533787 | 0.34 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr11_-_8892900 | 0.31 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr17_+_4675175 | 0.27 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr15_+_96875657 | 0.25 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr9_+_2158485 | 0.23 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr16_+_12059091 | 0.23 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_+_89300158 | 0.21 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_+_2158443 | 0.21 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_154377669 | 0.20 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr17_+_1665253 | 0.18 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_10687948 | 0.18 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_51897742 | 0.18 |
ENST00000600765.1
|
CTD-2616J11.14
|
CTD-2616J11.14 |
| chr17_-_27467418 | 0.17 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr17_+_76311791 | 0.16 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr3_-_112329110 | 0.16 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr22_-_42322795 | 0.15 |
ENST00000291232.3
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C |
| chr14_-_22005062 | 0.15 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr19_-_10687907 | 0.15 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_45973360 | 0.15 |
ENST00000589593.1
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr15_-_44069513 | 0.14 |
ENST00000433927.1
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr7_+_16793160 | 0.14 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr8_+_62200509 | 0.14 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr5_+_66254698 | 0.13 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr18_+_56532100 | 0.13 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr2_+_233390863 | 0.12 |
ENST00000449596.1
ENST00000543200.1 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr15_+_96876340 | 0.12 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_35856951 | 0.11 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr19_+_41281060 | 0.11 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr19_+_41281282 | 0.11 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr10_-_129924468 | 0.11 |
ENST00000368653.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr17_-_45056606 | 0.11 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr11_-_4629367 | 0.11 |
ENST00000533021.1
|
TRIM68
|
tripartite motif containing 68 |
| chr2_+_120687335 | 0.11 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr10_-_129924611 | 0.11 |
ENST00000368654.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr6_-_32920794 | 0.11 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr7_-_87104963 | 0.11 |
ENST00000359206.3
ENST00000358400.3 ENST00000265723.4 |
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr8_+_58055238 | 0.10 |
ENST00000519314.1
ENST00000519241.1 |
RP11-513O17.2
|
RP11-513O17.2 |
| chr4_-_467892 | 0.10 |
ENST00000506646.1
ENST00000505900.1 |
ZNF721
|
zinc finger protein 721 |
| chr5_+_148206156 | 0.10 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr11_-_125550726 | 0.10 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr11_+_34642656 | 0.10 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr11_-_4629388 | 0.10 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr16_+_50308028 | 0.10 |
ENST00000566761.2
|
ADCY7
|
adenylate cyclase 7 |
| chr17_+_68071389 | 0.10 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_-_123638633 | 0.10 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr2_-_89157161 | 0.10 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr14_-_83262540 | 0.10 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr4_-_185655212 | 0.10 |
ENST00000541971.1
|
MLF1IP
|
centromere protein U |
| chr11_+_1049862 | 0.10 |
ENST00000534584.1
|
RP13-870H17.3
|
RP13-870H17.3 |
| chrX_+_37639302 | 0.10 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_-_84121858 | 0.09 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_45908974 | 0.09 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr19_-_10687983 | 0.09 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr3_-_158450475 | 0.09 |
ENST00000237696.5
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr5_-_141338377 | 0.09 |
ENST00000510041.1
|
PCDH12
|
protocadherin 12 |
| chr2_+_108994466 | 0.09 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr2_-_142888573 | 0.09 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr1_-_54879140 | 0.09 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr1_-_114414316 | 0.09 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr19_+_36393367 | 0.08 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr12_-_50419177 | 0.08 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr17_-_26694979 | 0.08 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr7_-_130066571 | 0.08 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr2_-_204400113 | 0.08 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr9_-_95244781 | 0.08 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr4_-_144826682 | 0.08 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr1_-_109655377 | 0.08 |
ENST00000369948.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr1_-_179834311 | 0.08 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr11_-_64703354 | 0.08 |
ENST00000532246.1
ENST00000279168.2 |
GPHA2
|
glycoprotein hormone alpha 2 |
| chr18_-_46895066 | 0.08 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr18_-_30050395 | 0.08 |
ENST00000269209.6
ENST00000399218.4 |
GAREM
|
GRB2 associated, regulator of MAPK1 |
| chr5_+_111964133 | 0.08 |
ENST00000508879.1
ENST00000507565.1 |
RP11-159K7.2
|
RP11-159K7.2 |
| chr11_-_71823266 | 0.08 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr21_+_38593701 | 0.08 |
ENST00000440629.1
|
AP001432.14
|
AP001432.14 |
| chr1_-_110306562 | 0.08 |
ENST00000369805.3
|
EPS8L3
|
EPS8-like 3 |
| chr17_-_62340581 | 0.08 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr3_-_114343039 | 0.08 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_105332084 | 0.08 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr1_-_173793458 | 0.07 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr6_-_41673552 | 0.07 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr1_+_164528616 | 0.07 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr6_-_10412600 | 0.07 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr2_-_111291587 | 0.07 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr19_+_47813110 | 0.07 |
ENST00000355085.3
|
C5AR1
|
complement component 5a receptor 1 |
| chr17_-_39280419 | 0.07 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr11_+_20044375 | 0.07 |
ENST00000525322.1
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr1_-_109655355 | 0.07 |
ENST00000369945.3
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr17_-_56494908 | 0.07 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr4_+_48807155 | 0.07 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr11_-_8892464 | 0.07 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr2_-_231989808 | 0.07 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr17_-_18266818 | 0.07 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr2_+_11696464 | 0.07 |
ENST00000234142.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr21_+_17566643 | 0.07 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_+_53449887 | 0.07 |
ENST00000375327.3
|
RIBC1
|
RIB43A domain with coiled-coils 1 |
| chr7_+_114055052 | 0.07 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr19_+_55417499 | 0.07 |
ENST00000291890.4
ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr7_+_45067265 | 0.07 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr11_-_125550764 | 0.07 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr17_-_18266660 | 0.07 |
ENST00000582653.1
ENST00000352886.6 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr19_+_41281416 | 0.07 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr5_+_140174429 | 0.06 |
ENST00000520672.2
ENST00000378132.1 ENST00000526136.1 |
PCDHA2
|
protocadherin alpha 2 |
| chr17_+_37856253 | 0.06 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr16_+_2014941 | 0.06 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr17_-_57229155 | 0.06 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_+_46713404 | 0.06 |
ENST00000371975.4
ENST00000469835.1 |
RAD54L
|
RAD54-like (S. cerevisiae) |
| chr11_+_71900703 | 0.06 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr17_-_18266797 | 0.06 |
ENST00000316694.3
ENST00000539052.1 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr19_-_49140609 | 0.06 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_-_13900972 | 0.06 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr17_+_34171081 | 0.06 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr13_-_67802549 | 0.06 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr19_-_15311713 | 0.06 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
| chr3_+_63898275 | 0.06 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr9_-_75695323 | 0.06 |
ENST00000419959.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr17_-_56494882 | 0.06 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chrX_-_54069253 | 0.06 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr16_+_2014993 | 0.06 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr7_-_38948774 | 0.06 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr16_-_53737722 | 0.06 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr17_+_68071458 | 0.06 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr16_-_53737795 | 0.06 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr6_-_135271260 | 0.06 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr16_-_4838255 | 0.06 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr19_+_48958766 | 0.06 |
ENST00000342291.2
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr18_-_48723690 | 0.06 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C |
| chr1_+_153940741 | 0.06 |
ENST00000431292.1
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr2_-_99485825 | 0.06 |
ENST00000423771.1
|
KIAA1211L
|
KIAA1211-like |
| chr6_-_32908765 | 0.06 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr16_+_2533020 | 0.06 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr11_-_60720002 | 0.06 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr14_-_23285011 | 0.06 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_-_56494713 | 0.06 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr18_+_55712915 | 0.06 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_+_57828521 | 0.06 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr5_+_176784837 | 0.06 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr17_-_7018128 | 0.06 |
ENST00000380952.2
ENST00000254850.7 |
ASGR2
|
asialoglycoprotein receptor 2 |
| chr1_+_176432298 | 0.06 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr1_-_49242553 | 0.06 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr3_-_158450231 | 0.06 |
ENST00000479756.1
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr19_-_13227534 | 0.06 |
ENST00000588229.1
ENST00000357720.4 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr6_-_135271219 | 0.06 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr11_+_59522532 | 0.06 |
ENST00000337979.4
ENST00000535361.1 |
STX3
|
syntaxin 3 |
| chr18_+_52385068 | 0.06 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr7_-_92146729 | 0.06 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr11_-_104034827 | 0.05 |
ENST00000393158.2
|
PDGFD
|
platelet derived growth factor D |
| chr2_+_79412357 | 0.05 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr2_+_87755054 | 0.05 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr3_-_187388173 | 0.05 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr6_+_37225540 | 0.05 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr6_-_32908792 | 0.05 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_-_95623607 | 0.05 |
ENST00000531162.1
ENST00000529720.1 ENST00000343455.3 |
DICER1
|
dicer 1, ribonuclease type III |
| chr16_-_70323422 | 0.05 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chrX_-_41782683 | 0.05 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr17_+_37856214 | 0.05 |
ENST00000445658.2
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr10_-_82049424 | 0.05 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr19_-_44123734 | 0.05 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr19_-_13227463 | 0.05 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr1_-_161039753 | 0.05 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr12_-_59314246 | 0.05 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr17_-_26695013 | 0.05 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr7_+_151771377 | 0.05 |
ENST00000434507.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr21_-_36421535 | 0.05 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr12_-_106697974 | 0.05 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr12_+_54674482 | 0.05 |
ENST00000547708.1
ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr4_+_86749045 | 0.05 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_-_113627966 | 0.05 |
ENST00000505632.1
|
RP11-148B6.2
|
RP11-148B6.2 |
| chr3_+_149689066 | 0.05 |
ENST00000593416.1
|
AC117395.1
|
LOC646903 protein; Uncharacterized protein |
| chr17_-_7531121 | 0.05 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr17_-_39780819 | 0.05 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr1_+_61542922 | 0.05 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr7_-_1980128 | 0.05 |
ENST00000437877.1
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr15_+_91498089 | 0.05 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
| chr14_-_23285069 | 0.05 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr18_+_55711575 | 0.05 |
ENST00000356462.6
ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_71900572 | 0.05 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr12_-_58159361 | 0.05 |
ENST00000546567.1
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr6_-_31704282 | 0.05 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr17_+_26800296 | 0.05 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr11_+_118272328 | 0.05 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr17_-_18266765 | 0.05 |
ENST00000354098.3
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr6_+_45296391 | 0.05 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr19_-_18337240 | 0.05 |
ENST00000262805.12
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific |
| chr3_+_35680339 | 0.05 |
ENST00000450234.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_217082311 | 0.05 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chrX_+_103294483 | 0.05 |
ENST00000355016.3
|
H2BFM
|
H2B histone family, member M |
| chr3_-_114477962 | 0.05 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_+_104104379 | 0.05 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr15_+_42131011 | 0.05 |
ENST00000458483.1
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic) |
| chr1_+_165600436 | 0.05 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr4_+_41937131 | 0.05 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr9_+_2159850 | 0.05 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_+_60936921 | 0.05 |
ENST00000373878.3
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr2_+_26915584 | 0.05 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr19_-_44124019 | 0.05 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr21_-_36421401 | 0.05 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr1_+_173793777 | 0.05 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr18_-_78005231 | 0.05 |
ENST00000470488.2
ENST00000353265.3 |
PARD6G
|
par-6 family cell polarity regulator gamma |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.2 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0038185 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:1901846 | positive regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901846) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.0 | GO:1901657 | glycosyl compound metabolic process(GO:1901657) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.3 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.0 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |