Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for GAUAUGU
Z-value: 0.45
miRNA associated with seed GAUAUGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-190a-5p
|
MIMAT0000458 |
|
hsa-miR-190b
|
MIMAT0004929 |
Activity profile of GAUAUGU motif
Sorted Z-values of GAUAUGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23712312 | 0.31 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr11_-_31839488 | 0.28 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chr14_+_55034599 | 0.28 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr2_-_182545603 | 0.24 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr11_-_70507901 | 0.19 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_141645645 | 0.17 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr3_+_11314099 | 0.17 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr6_-_91006461 | 0.17 |
ENST00000257749.4
ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chrX_-_33146477 | 0.14 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr8_-_122653630 | 0.12 |
ENST00000303924.4
|
HAS2
|
hyaluronan synthase 2 |
| chr8_+_61591337 | 0.12 |
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7 |
| chr3_+_173116225 | 0.11 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_+_179059365 | 0.10 |
ENST00000190611.4
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr14_+_79745746 | 0.10 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr10_-_131762105 | 0.09 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr18_-_53255766 | 0.09 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr19_-_5340730 | 0.08 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr15_-_45815005 | 0.08 |
ENST00000261867.4
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr1_-_33283754 | 0.07 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr2_+_149402553 | 0.07 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr10_+_125425871 | 0.06 |
ENST00000284674.1
|
GPR26
|
G protein-coupled receptor 26 |
| chr16_+_24741013 | 0.06 |
ENST00000315183.7
ENST00000395799.3 |
TNRC6A
|
trinucleotide repeat containing 6A |
| chr13_+_49550015 | 0.05 |
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr19_-_33793430 | 0.05 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr11_-_30038490 | 0.05 |
ENST00000328224.6
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr18_-_35145593 | 0.05 |
ENST00000334919.5
ENST00000591282.1 ENST00000588597.1 |
CELF4
|
CUGBP, Elav-like family member 4 |
| chr6_+_73331776 | 0.04 |
ENST00000370398.1
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr1_+_145470504 | 0.04 |
ENST00000323397.4
|
ANKRD34A
|
ankyrin repeat domain 34A |
| chr3_-_126076264 | 0.03 |
ENST00000296233.3
|
KLF15
|
Kruppel-like factor 15 |
| chr11_-_106889250 | 0.03 |
ENST00000526355.2
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2 |
| chr7_+_5085452 | 0.03 |
ENST00000353796.3
ENST00000396912.1 ENST00000396904.2 |
RBAK
RBAK-RBAKDN
|
RB-associated KRAB zinc finger RBAK-RBAKDN readthrough |
| chr12_-_102874416 | 0.03 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr7_+_114055052 | 0.03 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr8_-_93115445 | 0.02 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_+_228336849 | 0.02 |
ENST00000409979.2
ENST00000310078.8 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr13_-_103053946 | 0.02 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chr15_-_52821247 | 0.02 |
ENST00000399231.3
ENST00000399233.2 |
MYO5A
|
myosin VA (heavy chain 12, myoxin) |
| chr22_+_40573921 | 0.02 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr20_+_56964169 | 0.01 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr7_-_26240357 | 0.01 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr2_+_208394616 | 0.00 |
ENST00000432329.2
ENST00000353267.3 ENST00000445803.1 |
CREB1
|
cAMP responsive element binding protein 1 |
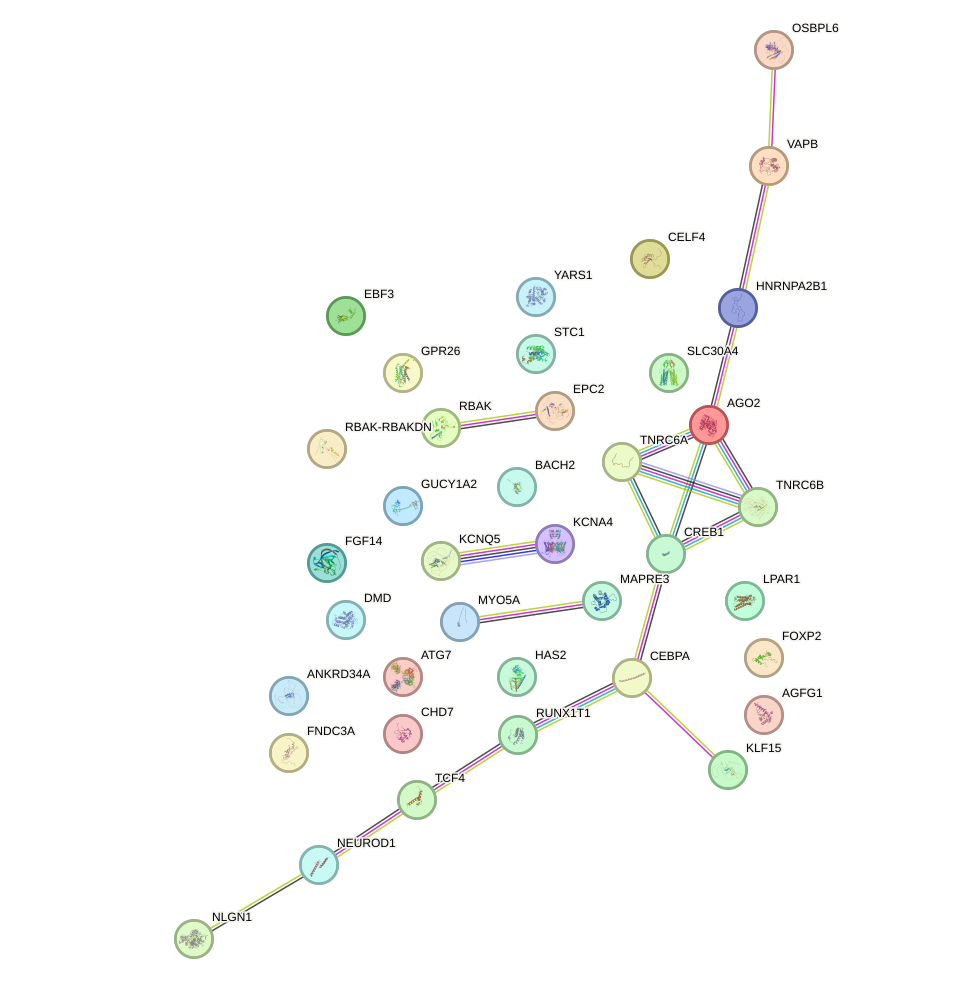
Network of associatons between targets according to the STRING database.
First level regulatory network of GAUAUGU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.2 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0021553 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |