Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
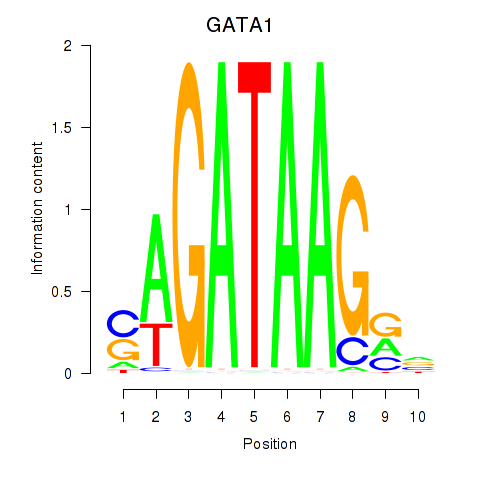
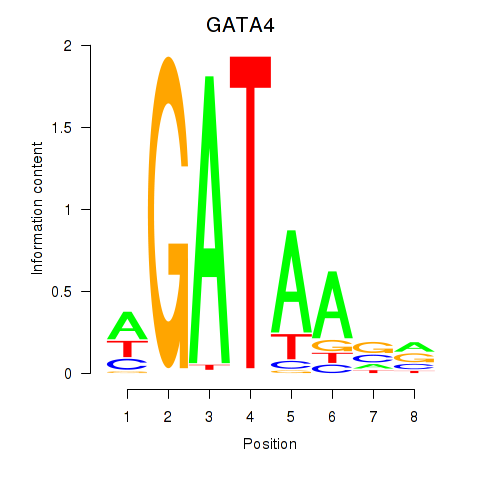
Results for GATA1_GATA4
Z-value: 0.45
Transcription factors associated with GATA1_GATA4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA1
|
ENSG00000102145.9 | GATA binding protein 1 |
|
GATA4
|
ENSG00000136574.13 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GATA4 | hg19_v2_chr8_+_11534462_11534475 | 0.87 | 1.3e-01 | Click! |
Activity profile of GATA1_GATA4 motif
Sorted Z-values of GATA1_GATA4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_235098935 | 0.33 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr17_+_4675175 | 0.33 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr1_-_235098861 | 0.31 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr19_-_39303576 | 0.24 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr1_-_226187013 | 0.23 |
ENST00000272091.7
|
SDE2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr17_-_64225508 | 0.23 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr8_+_107630340 | 0.21 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr11_-_10920838 | 0.19 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr14_+_88490894 | 0.19 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr19_-_4717835 | 0.18 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr17_-_42466864 | 0.17 |
ENST00000353281.4
ENST00000262407.5 |
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr10_-_99531709 | 0.17 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr19_-_51523275 | 0.16 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr6_-_107235331 | 0.16 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr17_+_46537697 | 0.16 |
ENST00000579972.1
|
RP11-433M22.2
|
RP11-433M22.2 |
| chr12_-_120765565 | 0.16 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr6_-_33663474 | 0.16 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr2_+_189156586 | 0.16 |
ENST00000409830.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr7_-_99277610 | 0.15 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr4_-_100140331 | 0.15 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr14_+_102276192 | 0.15 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr6_+_41010293 | 0.15 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr22_+_42665742 | 0.15 |
ENST00000332965.3
ENST00000415205.1 ENST00000446578.1 |
Z83851.3
|
Z83851.3 |
| chr16_+_333152 | 0.15 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr17_-_73840614 | 0.14 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr10_+_114710516 | 0.13 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr13_+_28527647 | 0.13 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr7_+_123469037 | 0.13 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr17_+_70036164 | 0.13 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr19_-_51522955 | 0.13 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr17_+_43224684 | 0.13 |
ENST00000332499.2
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1 |
| chr13_-_107214291 | 0.13 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr5_+_135496675 | 0.13 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr6_-_52628271 | 0.12 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr16_+_85936295 | 0.12 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr5_-_141703713 | 0.12 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr1_+_81106951 | 0.12 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr11_-_26743546 | 0.12 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr17_-_34079897 | 0.12 |
ENST00000254466.6
ENST00000587565.1 |
GAS2L2
|
growth arrest-specific 2 like 2 |
| chr2_-_14541060 | 0.12 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chrX_+_84258832 | 0.12 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr2_-_44065889 | 0.12 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr20_+_43029911 | 0.11 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr3_-_155394152 | 0.11 |
ENST00000494598.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr6_-_30128657 | 0.11 |
ENST00000449742.2
ENST00000376704.3 |
TRIM10
|
tripartite motif containing 10 |
| chr8_+_32579321 | 0.11 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr3_-_119379719 | 0.11 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr7_+_33765593 | 0.10 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr3_-_119379427 | 0.10 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr12_+_32655110 | 0.10 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_132479948 | 0.10 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr7_+_76139741 | 0.10 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr2_+_242289502 | 0.10 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr6_+_30614779 | 0.10 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr20_-_30795511 | 0.10 |
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2 |
| chr6_-_107235287 | 0.10 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr8_+_86999516 | 0.10 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chrX_+_152082969 | 0.09 |
ENST00000535861.1
ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185
|
zinc finger protein 185 (LIM domain) |
| chr10_+_180405 | 0.09 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr4_+_76871883 | 0.09 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr4_-_176733897 | 0.09 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr6_+_30614886 | 0.09 |
ENST00000376471.4
|
C6orf136
|
chromosome 6 open reading frame 136 |
| chr4_-_159094194 | 0.09 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr5_-_135290651 | 0.09 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr16_+_66442411 | 0.09 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr14_-_24584138 | 0.09 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr4_+_165675197 | 0.09 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr6_+_125540951 | 0.09 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr3_+_156807663 | 0.09 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr1_-_36020531 | 0.09 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr14_+_64680854 | 0.09 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr11_+_34642656 | 0.08 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr14_+_102276209 | 0.08 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr13_+_40229764 | 0.08 |
ENST00000416691.1
ENST00000422759.2 ENST00000455146.3 |
COG6
|
component of oligomeric golgi complex 6 |
| chrX_-_15402498 | 0.08 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr9_-_97356075 | 0.08 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr11_-_116708302 | 0.08 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr10_+_28822236 | 0.08 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr2_+_220436917 | 0.08 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr14_+_102276132 | 0.08 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_194207388 | 0.08 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr5_-_73937244 | 0.08 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr6_-_46889694 | 0.08 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr4_+_6910966 | 0.08 |
ENST00000444368.1
|
TBC1D14
|
TBC1 domain family, member 14 |
| chr1_-_238108575 | 0.08 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr3_-_155394099 | 0.08 |
ENST00000414191.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr5_-_141061777 | 0.08 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr14_+_53173890 | 0.08 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr12_-_91573132 | 0.08 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr2_-_38303218 | 0.08 |
ENST00000407341.1
ENST00000260630.3 |
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chrX_-_84634737 | 0.08 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr12_-_123634449 | 0.08 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr10_+_118349920 | 0.08 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr19_-_4066890 | 0.08 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr8_+_27629459 | 0.08 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr6_-_31846744 | 0.08 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr17_-_57229155 | 0.08 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr8_+_21823726 | 0.08 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr19_+_29704142 | 0.08 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr11_-_105892937 | 0.08 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr4_+_139694701 | 0.08 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr14_-_21566731 | 0.08 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr1_+_202172848 | 0.08 |
ENST00000255432.7
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr4_-_103266219 | 0.08 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr13_-_67804445 | 0.08 |
ENST00000456367.1
ENST00000377861.3 ENST00000544246.1 |
PCDH9
|
protocadherin 9 |
| chr4_+_187813673 | 0.07 |
ENST00000606881.1
|
RP11-11N5.3
|
RP11-11N5.3 |
| chr1_-_225616515 | 0.07 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr11_+_7595136 | 0.07 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_+_59783540 | 0.07 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_-_10601963 | 0.07 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr14_-_21945057 | 0.07 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr1_-_161207875 | 0.07 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr20_+_34713312 | 0.07 |
ENST00000373946.3
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr3_+_36421826 | 0.07 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr1_-_175162048 | 0.07 |
ENST00000444639.1
|
KIAA0040
|
KIAA0040 |
| chr19_+_37808778 | 0.07 |
ENST00000544914.1
ENST00000589188.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr11_-_125550726 | 0.07 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr12_+_104359576 | 0.07 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr17_-_79817091 | 0.07 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_+_150954493 | 0.07 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr14_+_24584372 | 0.07 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr16_+_71560154 | 0.07 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr18_+_61575200 | 0.07 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chrX_-_47930980 | 0.07 |
ENST00000442455.3
ENST00000428686.1 ENST00000276054.4 |
ZNF630
|
zinc finger protein 630 |
| chr2_-_187367356 | 0.07 |
ENST00000595956.1
|
AC018867.2
|
AC018867.2 |
| chr2_+_5832799 | 0.07 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr9_-_13175823 | 0.07 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_-_228028829 | 0.07 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr3_-_161090660 | 0.06 |
ENST00000359175.4
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr17_-_78194716 | 0.06 |
ENST00000576707.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr2_+_189156389 | 0.06 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_-_62084241 | 0.06 |
ENST00000449662.2
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr16_+_71560023 | 0.06 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr15_+_59279851 | 0.06 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr4_-_83821676 | 0.06 |
ENST00000355196.2
ENST00000507676.1 ENST00000506495.1 ENST00000507051.1 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr12_+_70219052 | 0.06 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr1_-_225615599 | 0.06 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chrX_+_78003204 | 0.06 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr4_+_74702214 | 0.06 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chrX_+_153030941 | 0.06 |
ENST00000538282.1
|
PLXNB3
|
plexin B3 |
| chr15_+_49913201 | 0.06 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr5_-_141061759 | 0.06 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr2_+_102927962 | 0.06 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr1_-_62190793 | 0.06 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr5_+_40841410 | 0.06 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr16_-_67978016 | 0.06 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr4_+_74269956 | 0.06 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr12_-_10320190 | 0.06 |
ENST00000543993.1
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr6_-_27840099 | 0.06 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr12_+_56477093 | 0.06 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr14_-_67981870 | 0.06 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr14_-_50559361 | 0.06 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr7_+_153584166 | 0.06 |
ENST00000404039.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr3_+_138340067 | 0.06 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr17_-_72864739 | 0.06 |
ENST00000579893.1
ENST00000544854.1 |
FDXR
|
ferredoxin reductase |
| chr7_-_27169801 | 0.06 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr1_+_161136180 | 0.05 |
ENST00000352210.5
ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX
|
protoporphyrinogen oxidase |
| chr20_+_53092232 | 0.05 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chr12_-_104359475 | 0.05 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr2_-_152830479 | 0.05 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_-_100815525 | 0.05 |
ENST00000226522.8
ENST00000499666.2 |
LAMTOR3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr7_-_99381884 | 0.05 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr8_-_30002179 | 0.05 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr4_+_83821835 | 0.05 |
ENST00000302236.5
|
THAP9
|
THAP domain containing 9 |
| chr5_+_68513622 | 0.05 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr8_+_124194752 | 0.05 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr3_-_93692781 | 0.05 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr14_-_65409438 | 0.05 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr18_+_42260861 | 0.05 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr3_-_178103144 | 0.05 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr3_-_120365866 | 0.05 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr1_+_174846570 | 0.05 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_-_54872059 | 0.05 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr8_+_99956759 | 0.05 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr19_-_51327034 | 0.05 |
ENST00000301420.2
ENST00000448701.2 |
KLK1
|
kallikrein 1 |
| chr10_-_63213177 | 0.05 |
ENST00000399298.3
ENST00000399293.1 |
TMEM26
|
transmembrane protein 26 |
| chr3_-_49726104 | 0.05 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr13_-_96329048 | 0.05 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr14_-_67981916 | 0.05 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chrX_-_72434628 | 0.05 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr21_-_34915123 | 0.05 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr16_-_71264558 | 0.05 |
ENST00000448089.2
ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN
|
HYDIN, axonemal central pair apparatus protein |
| chr6_-_138539627 | 0.05 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr3_+_52570610 | 0.05 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr15_+_78632666 | 0.05 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr15_-_52861323 | 0.05 |
ENST00000569723.1
ENST00000249822.4 ENST00000567669.1 ENST00000569281.2 ENST00000563566.1 ENST00000567830.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr9_-_124132483 | 0.05 |
ENST00000286713.2
ENST00000538954.1 ENST00000347359.2 |
STOM
|
stomatin |
| chr11_+_76571911 | 0.05 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr13_+_53029564 | 0.05 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr19_-_36822551 | 0.05 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chrX_+_38420623 | 0.05 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr6_+_130339710 | 0.05 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chrX_-_63615297 | 0.05 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr10_+_63808970 | 0.05 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr1_-_167487758 | 0.05 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr22_-_32651326 | 0.05 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr15_+_36887069 | 0.05 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr5_+_59783941 | 0.05 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr7_-_130066571 | 0.05 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr6_-_111927449 | 0.05 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GATA1_GATA4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.2 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) positive regulation of skeletal muscle tissue growth(GO:0048633) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.0 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0016250 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |